Gene Expression Changes Including Long Non-Coding RNAs in Human Hepatic Ischemia Reperfusion Injury
1Transplant Research Institute, James D. Eason Transplant Institute, Department of Surgery, The University of Tennessee Health Science Center, Memphis, TN, 2James D. Eason Transplant Institute, Department of Surgery, The University of Tennessee Health Science Center, Memphis, TN, 3Surgery, University of Maryland, Baltimore, MD, 4University of Maryland, Baltimore, MD, 5University of Tennessee/Methodist Transplant Institute, Memphis, TN
Meeting: 2022 American Transplant Congress
Abstract number: 660
Keywords: Gene expression, Ischemia
Topic: Basic Science » Basic Science » 14 - Ischemia Reperfusion
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:30pm-7:00pm
Presentation Time: 5:30pm-7:00pm
Location: Hynes Halls C & D
*Purpose: Ischemia reperfusion injury (IRI) is still one of the major problems in liver transplantation affecting long term outcomes. This study is sought out to understand the details of molecular mechanism for human hepatic IRI using next generation sequencing and human liver specimens.
*Methods: Pre-implantation and post reperfusion liver specimens were collected from liver transplant recipients (n=48). RNA was isolated from needle biopsies and bulk RNA-seq library was prepared. Pair-end sequencing was performed in NovaSeq. Reads were mapped to genome using STAR pipeline and differentially expressed genes in post reperfusion versus pre-implantation biopsies were accessed using DeSeq2 tool. KEGG analysis was performed to unravel the enriched pathways during human hepatic IRI.
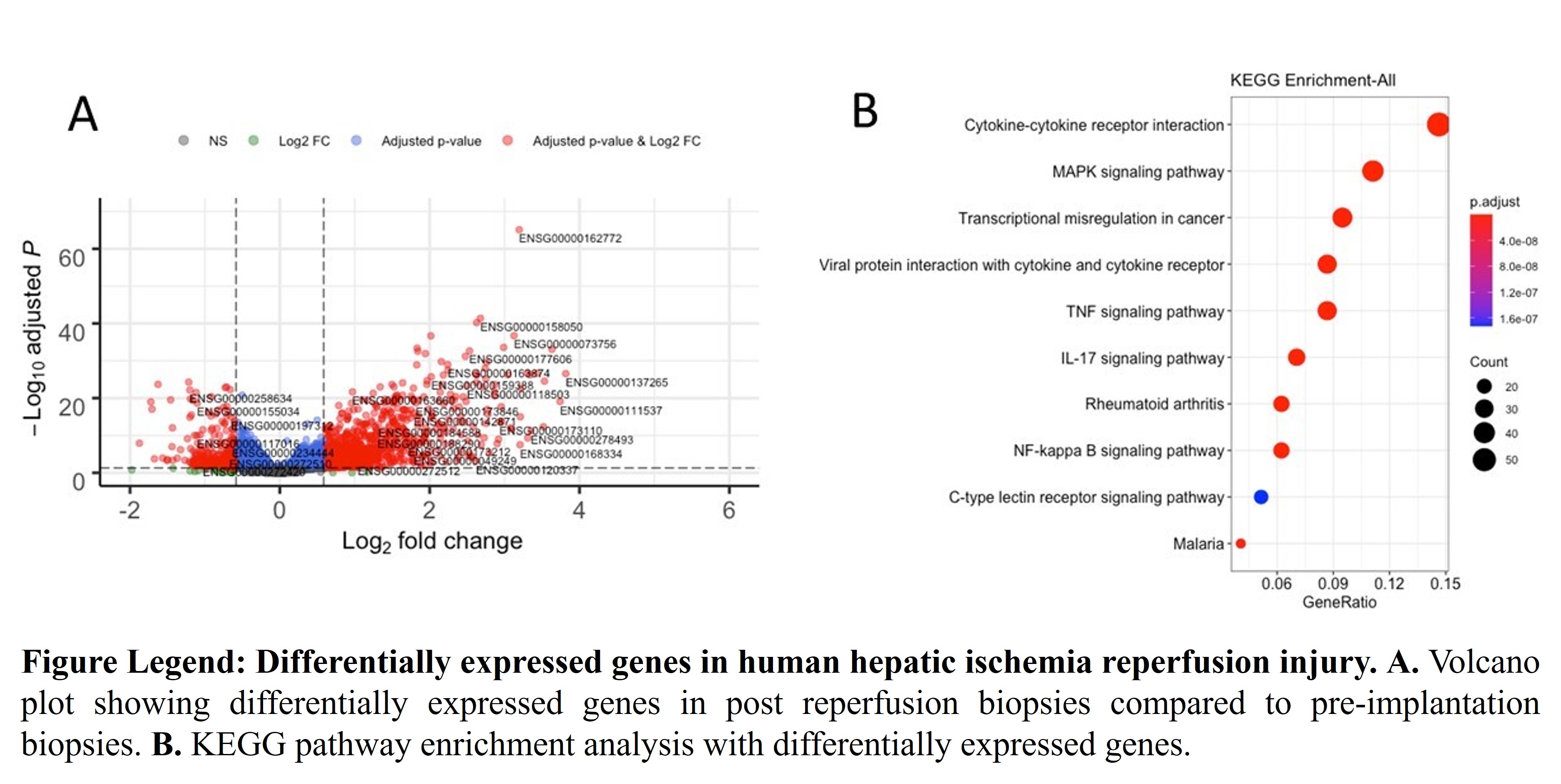
*Results: We identified 505 differentially expressed genes (439 overexpressed, 66 downregulated) with at least 2-fold changes and FDR < 0.05 in post reperfusion biopsies compared to pre-implantation biopsies (Fig. A). Interferon regulatory factor 4, interferon gamma, Cholesterol 25-hyroxylase, hydroxycarboxylic acid receptor 3, colony stimulating factor and FosB are among the top overexpressed genes indicating importance of immune and metabolic related changes. Pathway enrichment analysis showed cytokine-cytokine receptor interaction. MAPK, TNF, IL-17 and NF-kappa B signaling pathway (Fig. B). Most strikingly, 109 out of 505 differentially genes were long non-coding RNAs which were not implicated in human hepatic IRI in previous studies. Interestingly, 50 out of 66 downregulated genes are long-noncoding RNAs.
*Conclusions: Sequencing based profiling of gene expression changes using clinical needle biopsies is necessary to understand and resolve human hepatic IRI. This study is the first study reporting long non-coding RNA changes in liver IRI using clinical samples. Long non-coding RNAs are understudied and needs extra focus in human hepatic IRI.
To cite this abstract in AMA style:
Kuscu C, Naik S, Eymard C, Ingram H, Bajwa A, Maluf D, Mas V, Eason J, Kuscu C. Gene Expression Changes Including Long Non-Coding RNAs in Human Hepatic Ischemia Reperfusion Injury [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/gene-expression-changes-including-long-non-coding-rnas-in-human-hepatic-ischemia-reperfusion-injury/. Accessed February 23, 2026.« Back to 2022 American Transplant Congress