Discrepancy Rates Between Histology (HIST) and Molecular Diagnosis (MDX) of T-cell Mediated Rejection (TCMR) Depend on Choice of Bioinformatics Pipeline
University of Pittsburgh Medical Center, Pittsburgh, PA
Meeting: 2022 American Transplant Congress
Abstract number: 211
Keywords: Biopsy, Kidney, Rejection, T cells
Topic: Clinical Science » Kidney » 34 - Kidney: Acute Cellular Rejection
Session Information
Session Name: Kidney: Acute Cellular Rejection
Session Type: Rapid Fire Oral Abstract
Date: Monday, June 6, 2022
Session Time: 3:30pm-5:00pm
 Presentation Time: 4:10pm-4:20pm
Presentation Time: 4:10pm-4:20pm
Location: Hynes Ballroom A
*Purpose: HIST based treatment of TCMR has played a key role in the reduction of its incidence to historic lows of ~10%. MDX methods developed to enable more objective and quantitative assessment of TCMR injury show major discrepancies with HIST. This study was designed to assess the extent to which the choice of a bioinformatics pipeline can influence disagreement between HIST and MDX methods.
*Methods: HIST diagnoses were assigned to 45 renal allograft biopsies by a single pathologist to eliminate the effect of inter-observer reproducibility. Gene expression profiling was done by RNA-seq analysis of the entire biopsy core taken for HIST. Differential expression (DE) was performed on a selection holdout set of 10 TCMR and 10 STA (Stable) biopsies. Subsets of the 920 DE genes were used to train and test 5 different machine learning algorithms (MLs) for MDX of TCMR using 5-fold cross-validation on an independent 25-sample validation set.
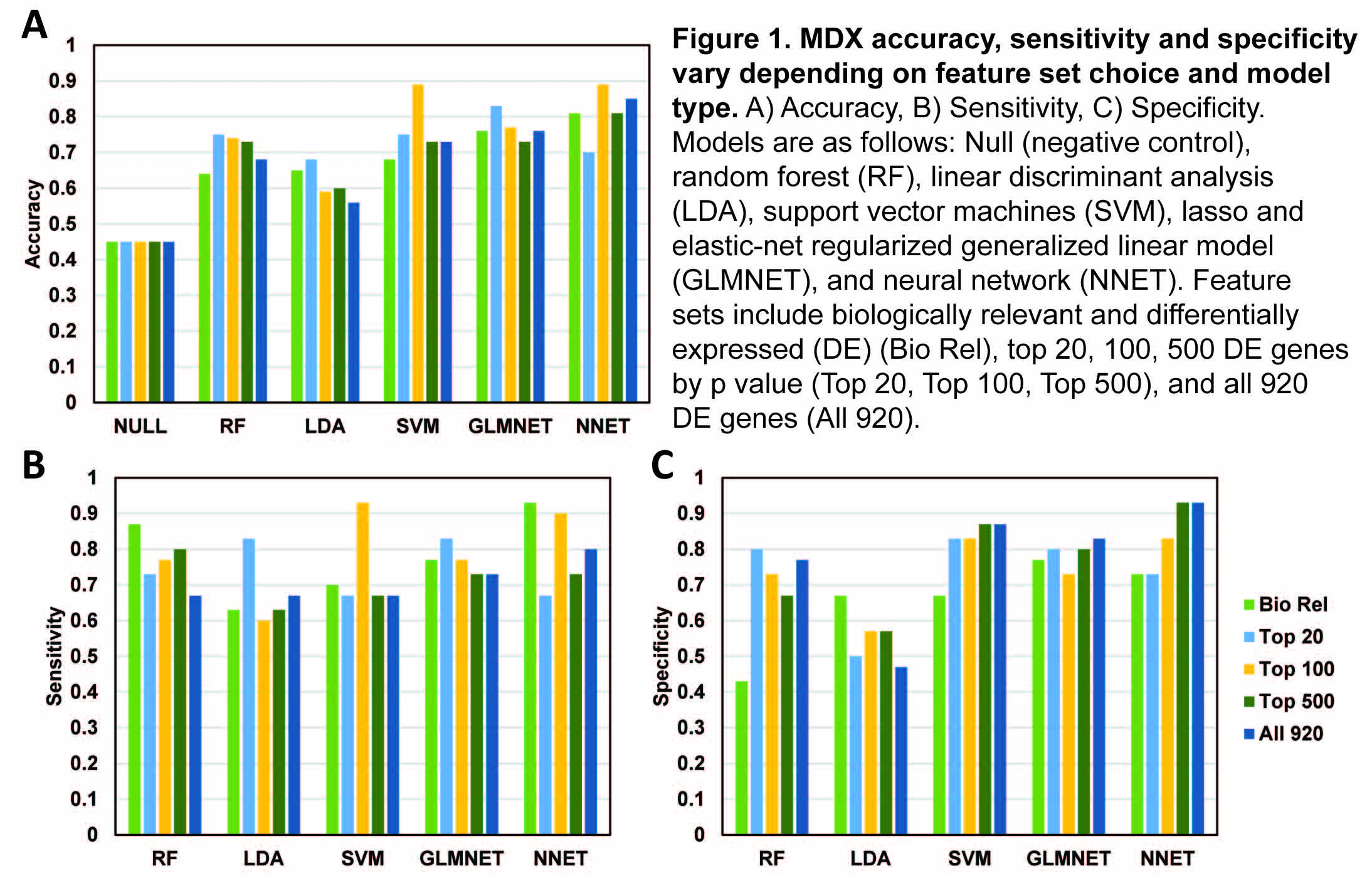
*Results: Based on the 920 DE genes, the diagnostic specificity for TCMR for the linear discriminant analysis (LDA), random forest (RF), GLMNET, support vector machines (SVM) and neural network (NNET) were respectively 0.47, 0.77, 0.83, 0.87 and 0.93 respectively. The corresponding sensitivities were 0.67, 0.67, 0.73, 0.67 and 0.80 respectively. The performance of NNET (the ML with highest overall accuracy) ranged between 0.70 and 0.85, as gene # was varied between 20 & 920. If MLs were, trained on the top 100 genes with the highest fold change, the accuracy of SVM improved from 0.73 to 0.89, and RF improved from 0.68 to 0.74. Training on DE genes selected from a pool of known biologically relevant genes improved sensitivity of RF (0.87) and NNET (0.93), but reduced their specificity (RF: 0.43, NNET: 0.73) and overall accuracy (RF: 0.64, NNET: 0.81). Notably, performance of LDA, the method used extensively in the development of the molecular microscope, was inferior to some of the other algorithms explored in this study.
*Conclusions: Labeling biopsies as TCMR or not-TCMR by MDX was significantly influenced by the bioinformatics pipeline used, with differences in specificity as high as 46%. Therefore, molecular assays must be clinically validated by histology-independent methods before they can be accepted as standard of care. Average values obtained from ensembles of multiple algorithms cannot be accepted as a representation of the ground truth, or as a means to correct histologic labels, since this average can itself vary with investigator choice of analytic method, with several hundred alternatives described in the literature.
To cite this abstract in AMA style:
Ellison M, Spagnolo D, Randhawa P. Discrepancy Rates Between Histology (HIST) and Molecular Diagnosis (MDX) of T-cell Mediated Rejection (TCMR) Depend on Choice of Bioinformatics Pipeline [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/discrepancy-rates-between-histology-hist-and-molecular-diagnosis-mdx-of-t-cell-mediated-rejection-tcmr-depend-on-choice-of-bioinformatics-pipeline/. Accessed February 21, 2026.« Back to 2022 American Transplant Congress