Comparative Analysis of Alloantigen-Stimulated Gene Expression in HIV(+) Liver Transplant Rejectors versus Non-Rejectors
Surgery, University of California, San Francisco, San Francisco, CA.
Meeting: 2018 American Transplant Congress
Abstract number: B6
Keywords: Genomic markers, HIV virus, Liver transplantation, Rejection
Session Information
Session Name: Poster Session B: Acute and Chronic Graft Injury
Session Type: Poster Session
Date: Sunday, June 3, 2018
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall 4EF
In a multi-center, prospective trial of solid-organ transplantation in persons with HIV, recipients had a 2-3x increased rejection rate when compared to historical data on HIV(-) recipients. Here, we perform alloantigen-stimulated gene expression profiling to investigate functional differences between HIV(+) rejectors and non-rejectors.
Methods: Donor and recipient specimens were collected prior to transplant and cryopreserved. Rejectors were selected based on biopsy-proven acute cellular rejection within 3 months of transplant. Non-rejectors were selected based on lack of clinical and biopsy-proven rejection. Propensity scores were matched between rejectors and non-rejectors. Third party was selected from a healthy donor with a high number of HLA mismatches. CD40L-stimulated donor or third party B cells were used to stimulate recipient cells in vitro prior to gene expression analysis using a customized NanoString panel. Raw counts were normalized and p-values were adjusted using the Benjamini-Hochberg procedure.
Results: Rejectors (n=2) showed high expression of T cell co-stimulators CD28 and ICOS. Interestingly, non-rejectors (n=2) exhibited high differential expression of genes associated with the complement system, including C1QA, C1QB, C2, CFB, and SERPING1. Moreover, the leukocyte immunoglobulin-like receptor (LILR) family of receptors were also upregulated in non-rejectors versus rejectors. Both donor and third party stimulation of recipient PBMCs resulted in similar gene expression profiles. 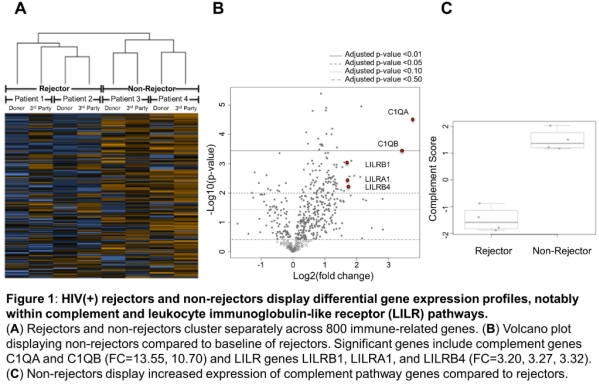
Conclusions: HIV(+) recipients with no rejection had higher expression of complement and LILR genes. These pathways have been implicated in facilitating immune cell tolerance. C1q is reported to play a fundamental role in facilitating apoptotic cell clearance and mediating immune suppression. LILRs have been shown to suppress T-cell activation via the induction of tolerogenic DCs. Similar profiles between donor and third party stimulation suggests that the differential gene expression is an intrinsic program of the recipient cells, not specific to a particular donor. Expanded analysis using this approach is warranted.
CITATION INFORMATION: Chu S., Lee K., Ward C., Stock P., Tang Q. Comparative Analysis of Alloantigen-Stimulated Gene Expression in HIV(+) Liver Transplant Rejectors versus Non-Rejectors Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Chu S, Lee K, Ward C, Stock P, Tang Q. Comparative Analysis of Alloantigen-Stimulated Gene Expression in HIV(+) Liver Transplant Rejectors versus Non-Rejectors [abstract]. https://atcmeetingabstracts.com/abstract/comparative-analysis-of-alloantigen-stimulated-gene-expression-in-hiv-liver-transplant-rejectors-versus-non-rejectors/. Accessed March 10, 2026.« Back to 2018 American Transplant Congress
