Characterization of CpG-stimulated, Tlr Regulatory, and Expanded Regulatory B Cells by RNA-Seq
Center for Transplant Science, MGH, Boston, MA
Meeting: 2020 American Transplant Congress
Abstract number: D-339
Keywords: B cells
Session Information
Session Name: Poster Session D: B-cell / Antibody /Autoimmunity
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Our group previously reported three different types of in vitro generated B cell populations. Expanded regulatory B cells (eBregs), TLR regulatory B cells (TLR-Bregs), and CpG-B cells have each been studied for their regulatory properties in vivo and in vitro. While their suppressive capabilities have been noted, differences in their transcriptomes have yet to be elucidated. This study aims to contrast transcriptional differences between three B cell expansion methods while highlighting certain differentially expressed genes (DEGs) as potential regulatory pathways.
*Methods: CpG-B cells, TLR-Bregs, and eBregs were obtained by giving isolated C57BL/6 splenic B cells either CpG ODN1668 stimulation for 3 days, CpG stimulation with LPS, PMA, and ionomycin over the last 5 hours, or an 8-day co-culture with irradiated CD40L-expressing NIH-3T3 cells containing BAFF, IL-4, anti-Tim-1, and IL-21. RNA was extracted from all groups and analyzed using RNA-Sequencing and R.
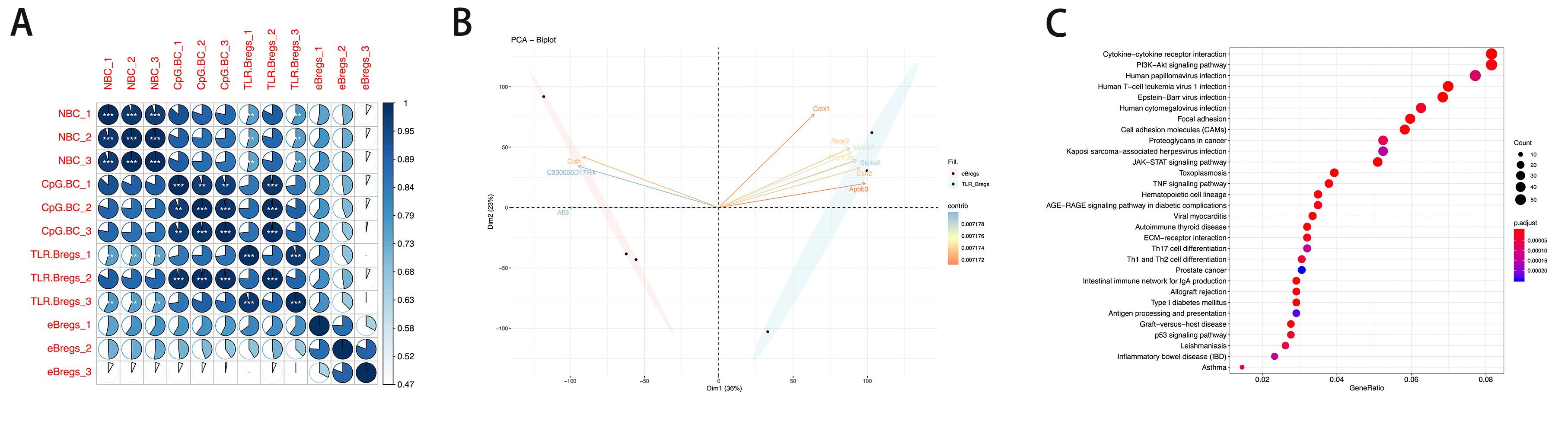
*Results: In comparison with Naïve B cell transcripts, there were 4178 DEGs (Log2|FC| > 1, P < 0.05) in the TLR-Breg group (n=3), of which 2001 transcripts were up-regulated and 2177 were down-regulated. Likewise, there were 4877 DEGs in the eBreg group (2776 up-regulated and 2101 down-regulated, n=3). Compared to TLR-Bregs, 1082 transcripts were up-regulated and 553 transcripts down-regulated in the eBreg group. Pearson correlation analysis showed that the characteristics among CpG-B cells, TLR-Bregs, and eBregs were significantly different (P < 0.05, Fig. A). According to PCA analysis, the 10 most altered transcripts were C330006D, Aff3, Slc4a2, Med17, Rcor2, Ppp1r3e, Cstf2, Ccbl1, Apbb3, Cish (Fig. B). KEGG analysis of the DEGs between TLR-Bregs and eBregs showed that the DEGs were largely enriched in allograft rejection, antigen processing and presentation, PI3K-Akt, TNF, Jak-STAT, and p53 signaling pathways (Fig. C).
*Conclusions: Though TLR-Bregs are derived from CPG-B cells, the DEGs indicate they are quite different. TLR-Bregs have been shown to inhibit naïve B cell proliferation while CPG-B cells do not. Transcriptome analyses among CPG-B cells, TLR-Bregs and eBregs provide potential areas for further research. These results offer insight into the underlying molecular mechanism behind regulatory B cell-induced tolerance.
To cite this abstract in AMA style:
Fu Q, Rickert GC, Lee K, Huai G, Deng K, Feeney N, Tanimine N, Cuenca A, Deng S, Markmann JF. Characterization of CpG-stimulated, Tlr Regulatory, and Expanded Regulatory B Cells by RNA-Seq [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/characterization-of-cpg-stimulated-tlr-regulatory-and-expanded-regulatory-b-cells-by-rna-seq/. Accessed February 19, 2026.« Back to 2020 American Transplant Congress