A Molecular Analysis of Graft Survival in the INTERHEART Study
1University of Alberta, Edmonton AB, AB, Canada, 2ATAGC, University of Alberta, Edmonton AB, AB, Canada
Meeting: 2019 American Transplant Congress
Abstract number: 58
Keywords: Gene expression, Heart, Survival
Session Information
Session Name: Concurrent Session: Biomarkers, Immune Monitoring and Outcomes I
Session Type: Concurrent Session
Date: Sunday, June 2, 2019
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:18pm-3:30pm
Presentation Time: 3:18pm-3:30pm
Location: Room 306
*Purpose: Rejection is a major cause of graft loss in heart and kidney transplants. The principal diagnosis associated with risk in kidneys is antibody-mediated rejection ‘ABMR’ (JCI Insight 2(12), 201710.1172/jci.insight.94197), and molecular rejection predicts graft failure better than histology (JASN 26(7):1711-1720, 2015). Similar comparisons in a heart transplant endomyocardial biopsy (EMB) population have not been performed.
*Methods: The INTERHEART study population contains 1265 indication and protocol EMB single bite biopsies from 18 centers in Canada, the USA, Australia and Europe. Affymetrix microarrays analyzed gene expression. 948 biopsies from 483 transplants (478 patients) had follow-up time and graft status. We selected 1 random biopsy per transplant and analyzed 3-year post-biopsy survival. Median follow-up time in this subset was 394 days, and 51/483 hearts failed by 3 years post-biopsy. We analyzed rejection by unsupervised archetype analysis using kidney-derived rejection-associated transcripts ‘RATs’ (JHLT 36:1192-1200, 2017) and by our interpretation of ISHLT histologic diagnosis.
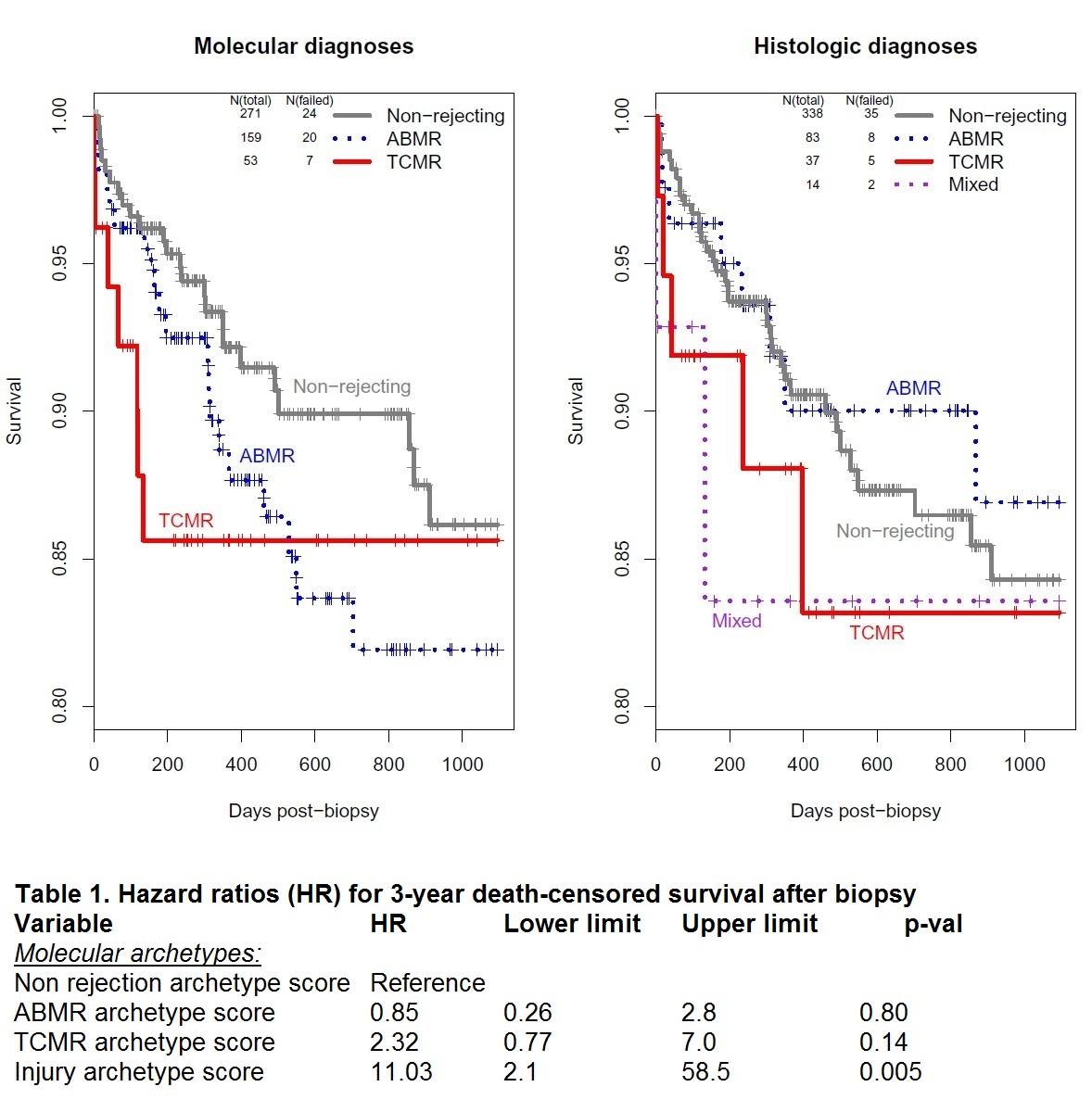
*Results: Four clusters of biopsies were found by archetypal analysis: 1) Non-rejection (N=686), 2) TCMR (129), 3) ABMR (437), and 4) Injury (13), used for the Kaplan-Meier plot of survival analysis in 483 transplants (Figure 1). Because only 4 hearts from cluster 4 remained (too few to analyze as a group), and 1 failed, each was incorporated into the next most closely associated archetype (3 moved to non-rejection, 1 of which failed, and 1 to TCMR). TCMR was a greater hazard than ABMR, both by molecules and histology (Figure 1). The results using histologic diagnoses were similar to those from molecular archetypes, except that the separation between ABMR and non-rejection was not as distinct using histology. Eleven biopsies, including one that failed, lacked a histologic diagnosis and were not included in the histologic survival analysis. In addition to belonging to a dominant archetype, each biopsy also has a score for each of the 4 archetypes, which permit the degree of molecular injury to be considered. These were used for multivariable Cox regression (Table 1). The only significant predictor of survival is degree of injury.
*Conclusions: In our study population, graft loss within the first years after EMB is more highly associated with TCMR than with ABMR. However, the best predictor of graft loss, as in kidneys, is the extent of injury regardless of its cause. Presumably TCMR (and ABMR) produce graft loss via molecular injury to the parenchyma. ClinicalTrials.gov # NCT02670408
To cite this abstract in AMA style:
Halloran P. A Molecular Analysis of Graft Survival in the INTERHEART Study [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/a-molecular-analysis-of-graft-survival-in-the-interheart-study/. Accessed February 26, 2026.« Back to 2019 American Transplant Congress