Evaluation of Genotype Based Clustering Method of Single-Cell RNA-Sequencing Data from Human Kidney Allograft Biopsies
1Weill Cornell Medicine, Doha, Qatar, 2Weill Cornell Medicine, New York, NY
Meeting: 2022 American Transplant Congress
Abstract number: 1311
Keywords: Biopsy, Genomics, Kidney transplantation, Rejection
Topic: Basic Science » Basic Science » 16 - Biomarkers: -omics and Systems Biology
Session Information
Session Name: Biomarkers: -omics and Systems Biology
Session Type: Poster Abstract
Date: Monday, June 6, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
*Purpose: Single-cell RNA sequencing (scRNA-seq) characterizes expression profiles of individual cells, providing novel insights into new cell types or new cell states that were not previously associated with clinical/pathological conditions. Allograft biopsies provide a unique cellular resource that contain cells of both recipient and donor origin. Identifying donor or recipient origin of cells is critical to further harness the benefits of scRNA-seq.
Herein, we evaluate the robustness of genotype-based deconvolution method of scRNA-seq data, in identifying the donor/recipient origin of cells.
*Methods: We selected scRNA-seq data from 3 human kidney allograft biopsies generated in our laboratory to evaluate the genotype-based deconvolution method; 2 were donor/recipient sex mismatched and 1 was obtained at the time of donor nephrectomy. We used 10x Genomics Cell Ranger 6.1.1 to individually align raw scRNA-Seq data to human genome reference (GRCh38) downloaded from 10x Genomics with default parameters. The output BAM and barcode file were used as inputs for singularity version of Souporcell pipeline. The cluster data was merged into Seurat object in R version 4.1.0. The plot was generated using ggplot2 package.
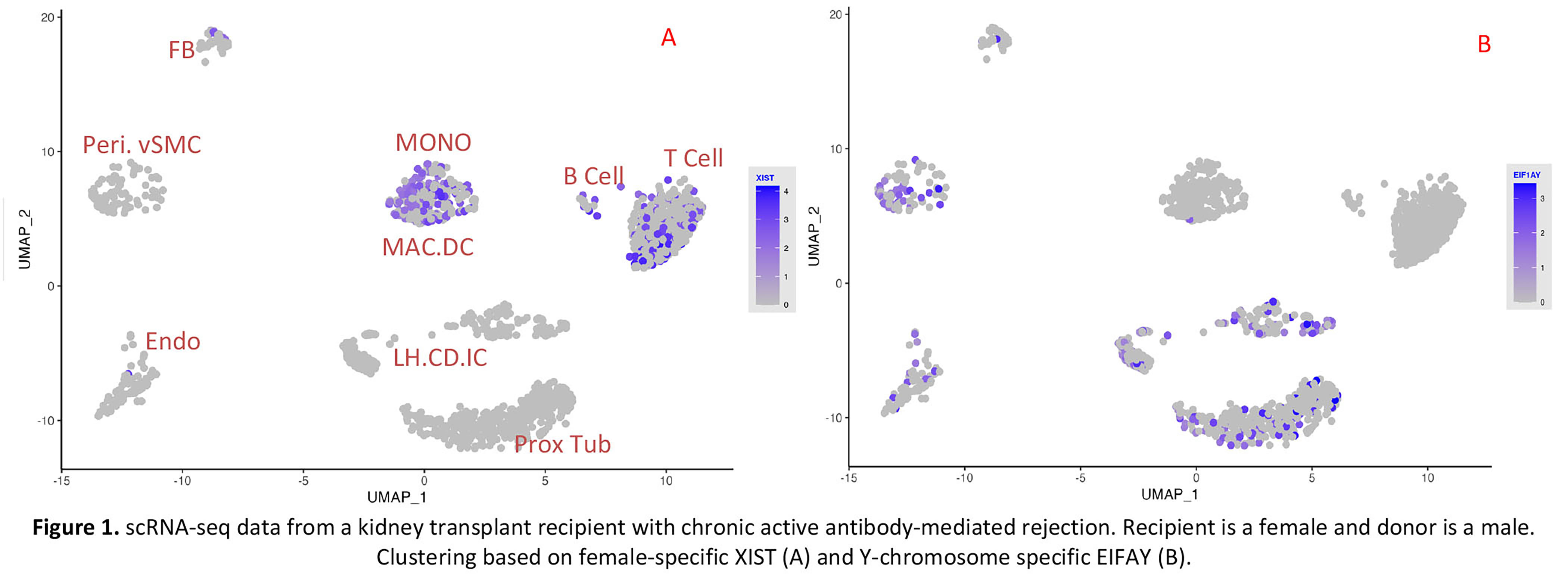
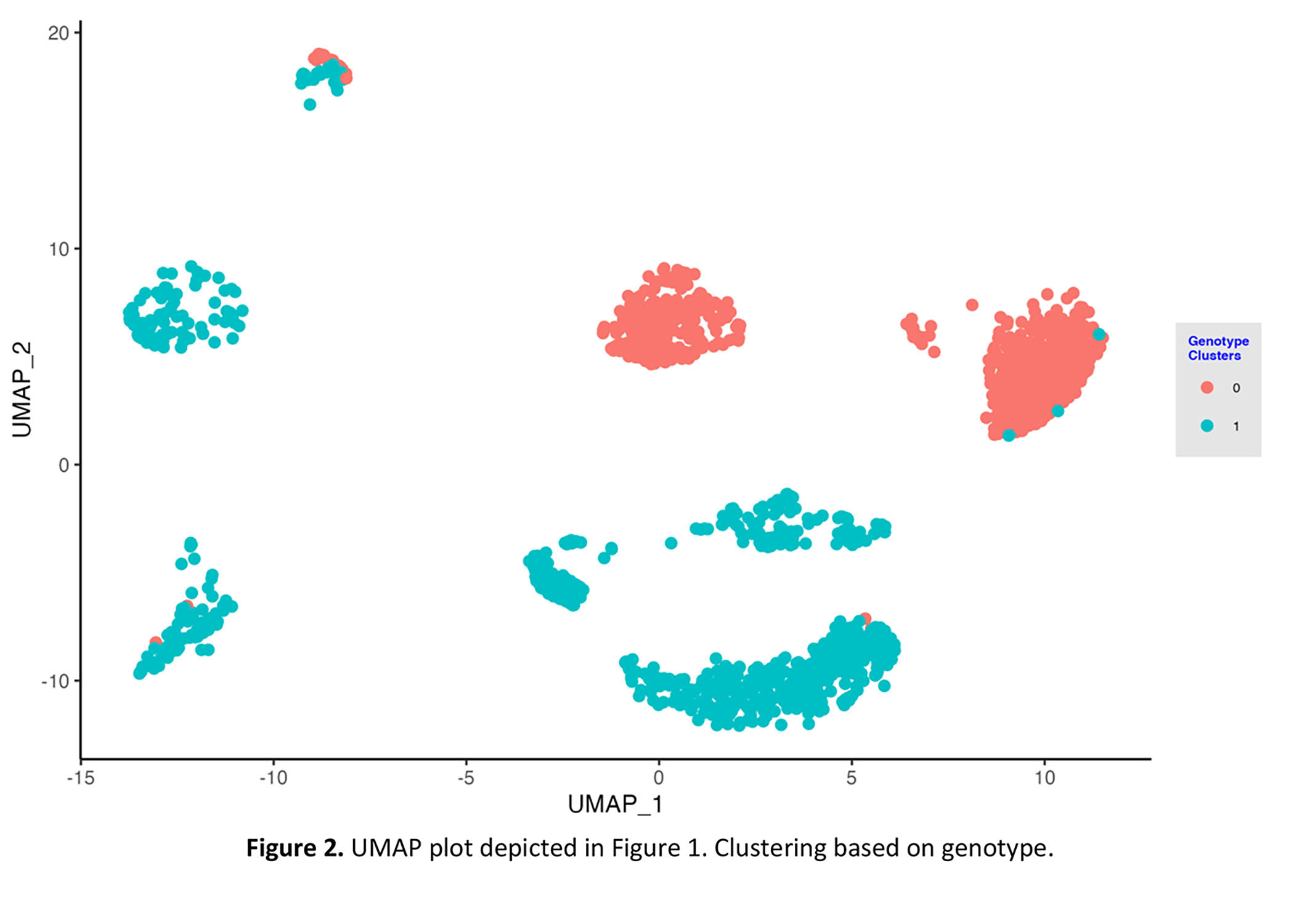
*Results: We first used the expression of female-specific marker XIST and Y-chromosome marker, EIF1Y, to cluster the donor/recipient origin of cells. We next applied the Souporcell pipeline. The genotype-based clustering was similar to the sex-matched clustering. Importantly, donor genotype information is not needed for identifying the donor or recipient origin of the cells.
A representative example of one biopsy in which the recipient was a female and the donor was a male is shown (Figure 1 and 2).
*Conclusions: Our successful application of genotype-based deconvolution method to determine the donor/recipient origin of the cells in scRNA-seq of human kidney allograft biopsies will further our ability to study cellular interactions between donor and recipient cells during rejection.
To cite this abstract in AMA style:
Thareja G, Belkadi A, Rahim S, Yang H, Mueller FB, Suhre K, Suthanthiran M, Muthukumar T. Evaluation of Genotype Based Clustering Method of Single-Cell RNA-Sequencing Data from Human Kidney Allograft Biopsies [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/evaluation-of-genotype-based-clustering-method-of-single-cell-rna-sequencing-data-from-human-kidney-allograft-biopsies/. Accessed March 3, 2026.« Back to 2022 American Transplant Congress