Absolute Quantification of Donor-Derived Cell-Free DNA Does Not Provide Additional Discriminating Power Over Donor-derived Cell-Free DNA Fraction for Detection of Allograft Rejection
1Alpert School of Medicine Brown University, Providence, RI, 2Cedars Sinai Medical Ctr, Los Angeles, CA, 3CareDx, Redwood City, CA, 4University of Maryland School of Medicine, Baltimore, MD
Meeting: 2022 American Transplant Congress
Abstract number: 1394
Keywords: Biopsy, Graft function, Kidney transplantation, Monitoring
Topic: Clinical Science » Kidney » 41 - Kidney Technical
Session Information
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:30pm-7:00pm
Presentation Time: 5:30pm-7:00pm
Location: Hynes Hall C
*Purpose: High level of background cell-free DNA (cfDNA) has been hypothesized as a concern for false negative results in molecular diagnostics for allograft rejection that rely on fraction of donor derived-cfDNA (dd-cfDNA), generating interest in the use of absolute quantification (AbQ) of dd-cfDNA. Here we assessed the value of total cfDNA and AbQ dd-cfDNA in comparison to fraction of dd-cfDNA for the detection of allograft rejection.
*Methods: 214 samples from 164 patients from the Kidney Allograft Outcomes AlloSure Registry (KOAR) were assessed for total cfDNA, AbQ of dd-cfDNA, and fraction of dd-cfDNA within 30 days of biopsy. The dataset represented the subset of the KOAR samples, for which total cfDNA, AbQ, AlloSure, and biopsy were able to be obtained. It included 150 non-rejections (normal) and 64 rejections; acute AMBR (n=11), acute TCMR 1A (n=20), acute TCMR 1B (n=7), acute TCMR 2B (n=1), chronic active ABMR (n=5), chronic active TCMR (n=5), suspicious ABMR (n=6) and mixed (n=9). Total DNA and cfDNA was quantified using Tapestation 4200. AbQ (copies per ml) was calculated using a proprietary method and algorithm. Fraction of dd-cfDNA was performed using AlloSure. The 3 metrics were independently tested for their ability to discriminate rejections vs non-rejections. The data were then further randomly divided into a training (70%) and test (30%) set to evaluate performance of a combined score.
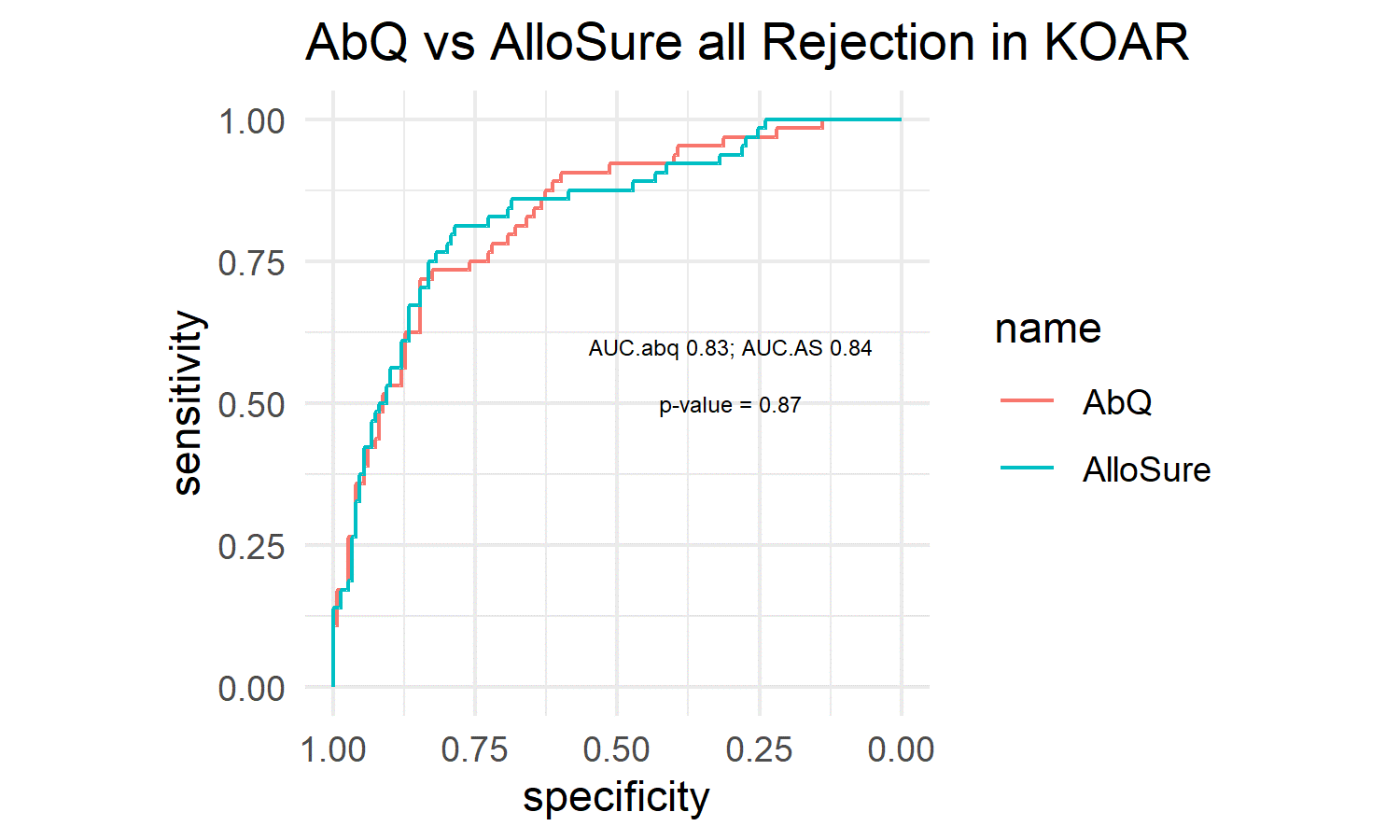
*Results: Total cfDNA provided no discrimination between rejections and non-rejections. The AUCs for predicting rejection using AbQ and AlloSure were not significantly different from one another (p=0.87), with AUC.AbQ=0.83, AUC.AlloSure=0.84.
Using the most optimal threshold for both metrics, performance was similar. For fraction dd-cfDNA, sensitivity = 0.81, specificity = 0.79, PPV = 0.62%, NPV = 0.91. For AbQ, sensitivity = 0.72, specificity = 0.85, PPV = 0.67, NPV = 0.88. When both metrics were combined, performance was not statistically any better than either metric alone, with AUCs being 0.85 in both the training and test set.
*Conclusions: AbQ of dd-cfDNA does not provide any additional discriminating power over dd-cfDNA fraction for detection of allograft rejection.
To cite this abstract in AMA style:
Gohh R, Jordan S, Woodward RN, Clark-Langone KM, Morlan J, Silva B, Weir M. Absolute Quantification of Donor-Derived Cell-Free DNA Does Not Provide Additional Discriminating Power Over Donor-derived Cell-Free DNA Fraction for Detection of Allograft Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/absolute-quantification-of-donor-derived-cell-free-dna-does-not-provide-additional-discriminating-power-over-donor-derived-cell-free-dna-fraction-for-detection-of-allograft-rejection/. Accessed February 17, 2026.« Back to 2022 American Transplant Congress