Deep Phenotyping Methods Highlight Recipient-Derived CD16+ NK and CD16+ Monocyte Infiltration During Kidney Transplant Rejection
1Microbiology, Immunology and Transplantation, KU Leuven, Leuven, Belgium, 2Department of Morphology and Molecular Pathology, University Hospitals Leuven, Leuven, Belgium, 3Faculty of Medicine, KU Leuven, Leuven, Belgium, 4Laboratory for Translational Genetics, Department of Human Genetics, KU Leuven, Leuven, Belgium, 5ESAT Stadius Center for Dynamical Systems, Signal Processing and Data Analytics, KU Leuven, Leuven, Belgium
Meeting: 2022 American Transplant Congress
Abstract number: 376
Keywords: Biopsy, Gene expression, Kidney transplantation, Rejection
Topic: Basic Science » Basic Science » 02 - Acute Rejection
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 6:40pm-6:50pm
Presentation Time: 6:40pm-6:50pm
Location: Hynes Room 309
*Purpose: How the immune system participates to kidney transplant rejection has not been fully elucidated. Single-cell (sc) genomics techniques are revolutionizing our ability to characterize complex disease processes such as rejection, coupled with the ability to examine transcriptional profiles at single-cell resolution.
*Methods: Here, we used sc-RNA sequencing (10x Genomics platform) to define the global immune landscape of human kidney transplants. We studied sc-suspensions (N= 35,152 cells) from 16 different allograft biopsies, with a representative mix of clinicopathological phenotypes (3 antibody-mediated rejection (ABMR), 1 mixed rejection, 8 borderline changes, and 4 stable cases).
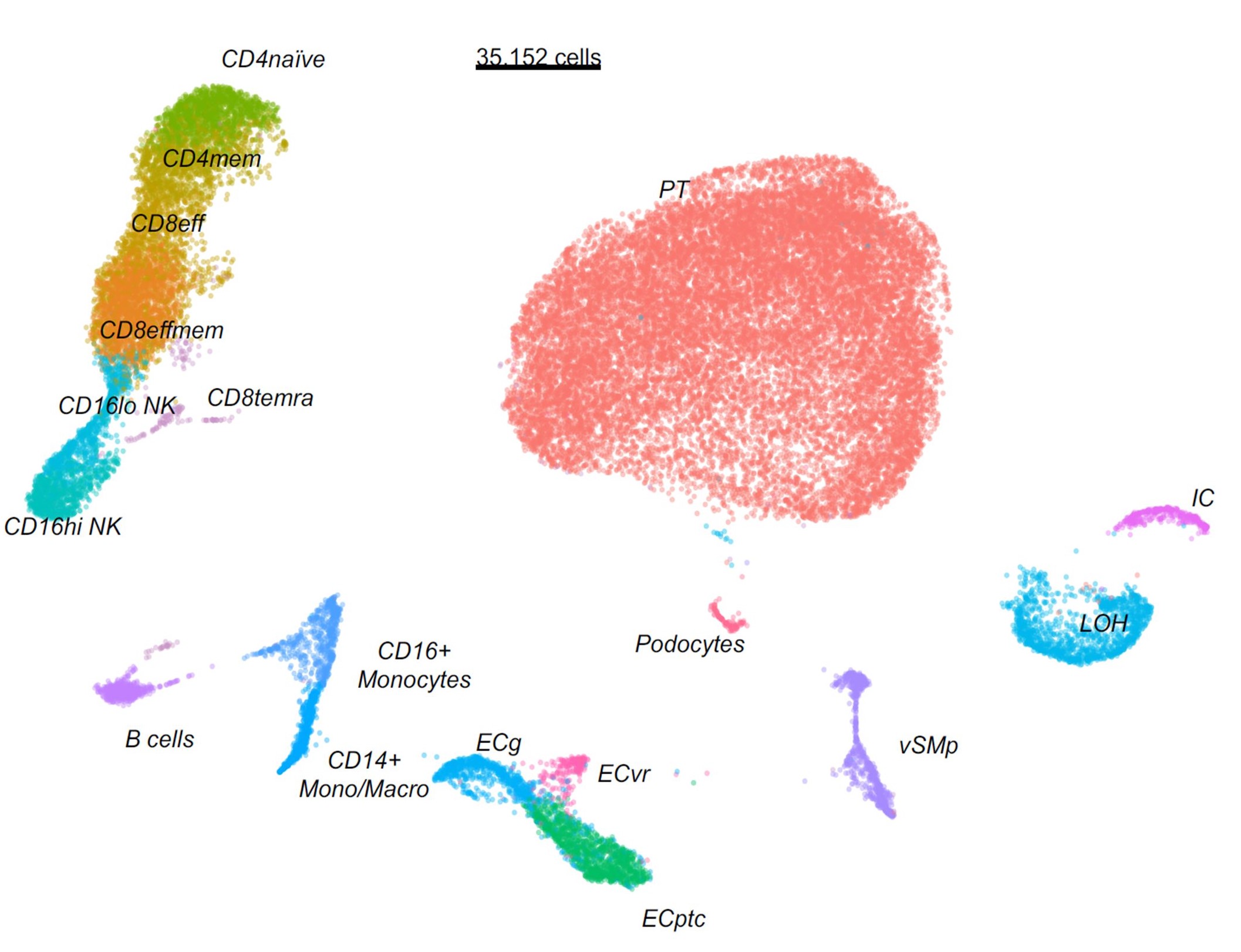
*Results: We identified 18 distinct cell types, including all major immune cell types and most kidney cell types. Nephron epithelial cells were evident, such as podocytes, peritubular cells (PT), vascular smooth muscle and pericytes (vSMp) and Loop of Henle (LOH) cells. We also observed different clusters of endothelial cells (EC) such as glomerular EC (ECg), vasa recta EC (ECvr) and peritubular capillaries (ECptc). Immune cell populations included CD16+ monocytes, CD14+ monocytes and macrophages, B cells, different CD4 and CD8 T cells, and two subsets of Natural Killer (NK) cells: CD16hi and CD16lo NK cells. Using a data-driven phenotypic stratification of biopsies with a high histological and clinical relevance, we observed an association of CD16+ NK and monocytes infiltration with inflammation severity. Ligand-receptor analysis revealed that CD16hiNK cells and monocytes relied essentially on shared incoming signals such as MIF, SPP1, CXCL and TNF but their outgoing signaling differed. Outgoing NK cell signaling was characterized by CCL pathways whereas outgoing CD16+ monocytes signaling was driven by TNF, CXCL and MIF pathways.
*Conclusions: Our study provides new insights of the role of the different innate immune cell populations and their interactions with the kidney and immune cells and unravels CD16+ NK cells and monocytes as key-players during kidney rejection.
To cite this abstract in AMA style:
Lamarthée B, Callemeyn J, Koshy P, Debyser T, Loon EVan, Franken A, Brussel TVan, Arijs I, Lambrechts D, Senev A, Vaulet T, Tinel C, Naesens M. Deep Phenotyping Methods Highlight Recipient-Derived CD16+ NK and CD16+ Monocyte Infiltration During Kidney Transplant Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/deep-phenotyping-methods-highlight-recipient-derived-cd16-nk-and-cd16-monocyte-infiltration-during-kidney-transplant-rejection/. Accessed January 30, 2026.« Back to 2022 American Transplant Congress