Digital Spatial Profiling of Kidney Allograft Biopsies with Chronic Allograft Dysfunction in African American Transplant Recipients
1Hennepin Healthcare Research Institute, Minneapolis, MN, 2University of Minnesota, Minneapolis, MN, 3Hennepin County Medical Center, Minneapolis, MN, 4Medicine, University of Nebraska Medical Center, Omaha, NE, 5University of Alabama at Birmingham, Birmingham, AL, 6Stanford University, Palo Alto, CA
Meeting: 2022 American Transplant Congress
Abstract number: 114
Keywords: African-American, Biopsy, Graft failure, Kidney transplantation
Topic: Clinical Science » Kidney » 47 - Kidney Complications: Immune Mediated Late Graft Failure
Session Information
Session Name: Kidney Complications: Chronic Antibody Mediated Rejection & Immune Mediated Late Graft Failure
Session Type: Rapid Fire Oral Abstract
Date: Sunday, June 5, 2022
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:40pm-5:50pm
Presentation Time: 5:40pm-5:50pm
Location: Hynes Room 302
*Purpose: Understanding tissue heterogeneity is important to identify the molecular and cellular mechanisms of kidney allograft loss in African American (AA) transplant recipients. Our aim is to use digital spatial profiling (DSP) to identify the presence of different cell populations, transcripts and their locations associated with allograft loss.
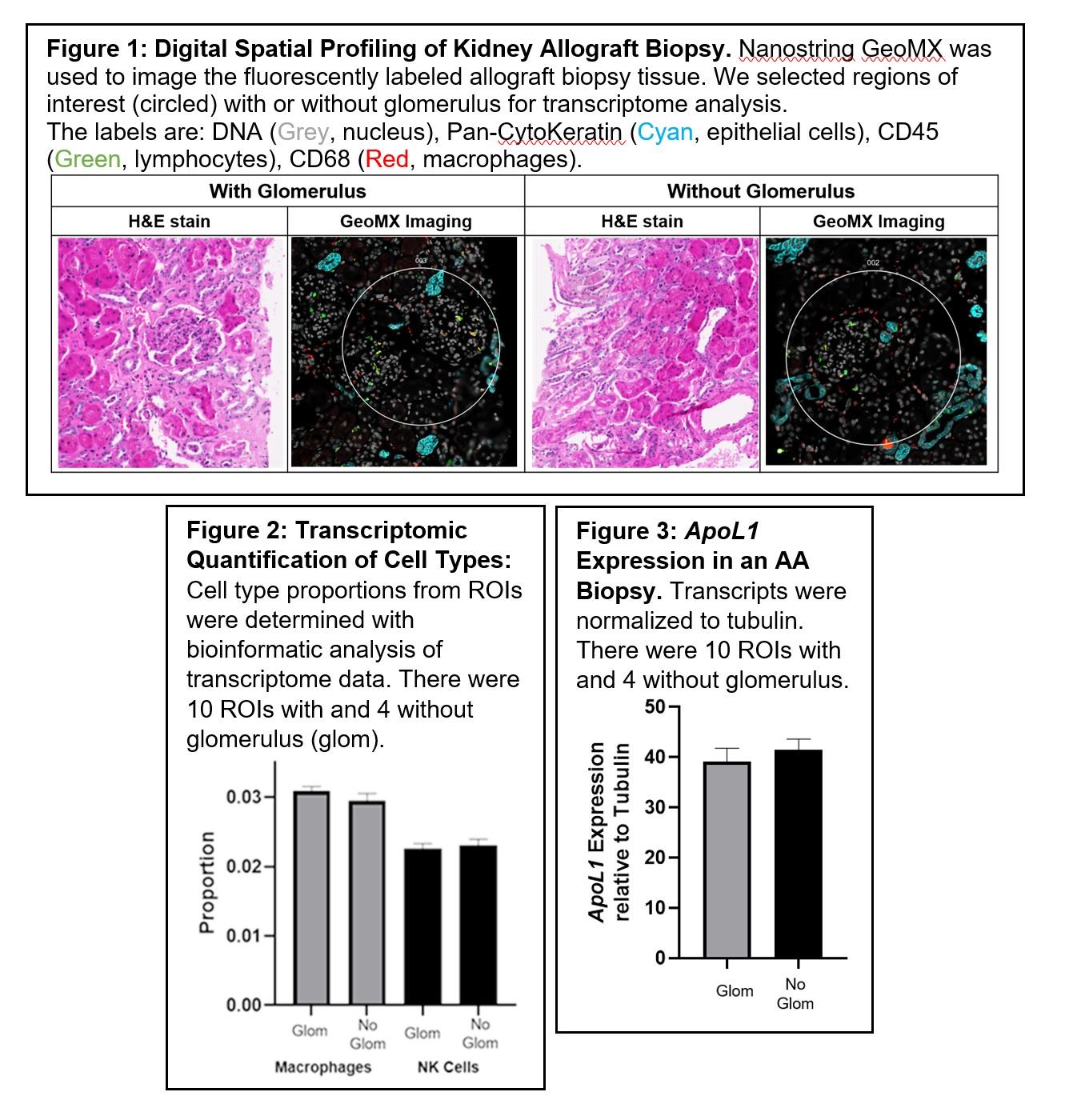
*Methods: AA patients, with chronic allograft dysfunction (CGD), enrolled in the Deterioration of Kidney Allograft Function (DeKAF) study, with or without eventual allograft loss, were included in this study. CGD was defined as a >25% increase in creatinine relative to a baseline at 3 months post-transplant triggering a kidney biopsy. Formalin-fixed paraffin-embedded (FFPE) kidney biopsy tissues from recipients were assessed by Nanostring GeoMX DSP after antibody staining for pan-cytokeratin, CD45, CD68, and Syto-13, to identify specific cell populations. Nanostring’s Whole Transcriptome Atlas (WTA) sequencing was used to quantify the each transcript. Up to 14 regions of interest (ROIs) were selected per biopsy. Histology showed more fibrosis in the interstitial and tubular regions of the biopsies in AAs so we separated the ROIs with or without glomerulus (interstitial/tubular regions). SingleR analysis with the human cell atlas annotation reference was used to deconvolute and quantify the cell types from transcriptome data.
*Results: Figure 1 shows the H&E and fluorescent images of an AA biopsy without allograft loss. We were able to identify mononuclear cells in biopsy such as natural killer (NK) cells and macrophages. Figure 2 shows that we can quantify cell types in different locations of the the biopsy from whole transcriptome data. Since ApoL1 variants, enriched in AA donor kidneys, are risk factors for poor outcomes in recipients, Figure 3 shows that we can quantify the Apol1 expression in kidney allografts.
*Conclusions: DSP allows for the identification, localization and quantification of specific cell types (macrophages and NK cells) and molecular transcripts in kidney allograft biopsies. These data can be used for association studies to determine biological mechanisms of allograft loss in AA transplant recipients.
To cite this abstract in AMA style:
Dorr CR, Elmer SA, Wu B, Guo B, Czyzyk J, Crary G, Onyeaghala G, Oetting W, Jacobson P, Mannon RB, Agarwal G, Maltzman J, Matas A, Israni A. Digital Spatial Profiling of Kidney Allograft Biopsies with Chronic Allograft Dysfunction in African American Transplant Recipients [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/digital-spatial-profiling-of-kidney-allograft-biopsies-with-chronic-allograft-dysfunction-in-african-american-transplant-recipients/. Accessed February 9, 2026.« Back to 2022 American Transplant Congress