Single Cell RNAq-seq Analysis of Paired Peripheral and Intragraft T Cells Reveals Biopsy Specific Fc Receptor Gene Expression in Rejection Only
Washington University School of Medicine, St Louis, MO
Meeting: 2021 American Transplant Congress
Abstract number: 564
Keywords: Gene expression, Kidney transplantation, T cells
Topic: Basic Science » Lymphocyte Biology: Signaling, Co-Stimulation, Regulation
Session Information
Session Name: Lymphocyte Biology: Signaling, Co-Stimulation, Regulation
Session Type: Poster Abstract
Session Date & Time: None. Available on demand.
Location: Virtual
*Purpose: Antibody mediated rejection (AMR) remains one of the major causes of allograft failure and our understanding of this disease process is poor. Transcriptomics studies have suggested antibody binding to Fc receptors plays a role in allograft injury by triggering T cell and NK cell activation in AMR. We performed single cell RNAseq on paired peripheral blood and biopsies from patients with AMR and non-rejection focusing on T cells and Fc receptor expression.
*Methods: The 10X Genomics platform was used to make libraries which were sequenced to a depth of ~50k reads/cell. Gene-cell matrices were obtained from CellRanger and the downstream analysis (clustering, integration analysis, expression analyses) were done using R, Seurat and Monocle2. This study had IRB approval.
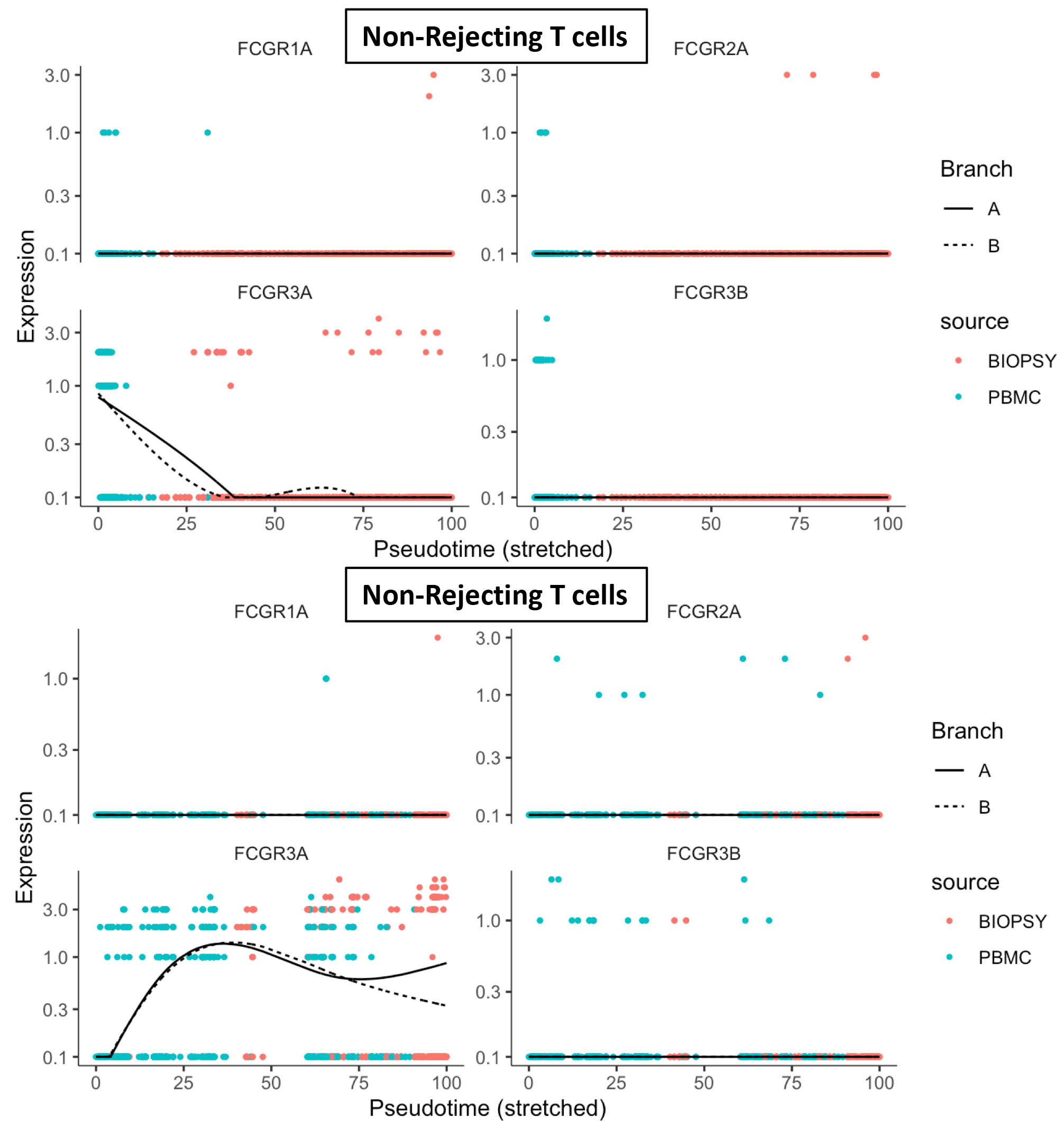
*Results: 16086 immune cells in total (avg = 1124 genes/cell) from 5 kidney transplant patients (biopsy and pbmc’s) were included in the final integrated analysis using UMAP. All major kidney cell types were identified including macrophages, B cells and T cells. T cells from rejecting samples (2442 T cells) and non-rejecting samples (2971 T cells) were subset for further analysis. A pseudotime analysis using Monocle2 was performed on these T cell subsets. We examined the expression of the Fc receptor genes, FCGR3B, FCGR2A, FCGR3A, FCGR1A, FCGR2C. We found that only FCGR3A (CD16a) varied with pseudotime and that expression increased from peripheral T cells to intragraft T cells in AMR patients only.
*Conclusions: Our study find that FCGR3A expression is increased in T cells in AMR and is specific for the local environment of the allograft. These data confirm intragraft T cells as the source FCGR3A expression previously suggested in microarray based studies.
To cite this abstract in AMA style:
Malone A, Leckie-Harre A, Silverman I, Chadha A, Wu H, Humphreys B. Single Cell RNAq-seq Analysis of Paired Peripheral and Intragraft T Cells Reveals Biopsy Specific Fc Receptor Gene Expression in Rejection Only [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/single-cell-rnaq-seq-analysis-of-paired-peripheral-and-intragraft-t-cells-reveals-biopsy-specific-fc-receptor-gene-expression-in-rejection-only/. Accessed February 19, 2026.« Back to 2021 American Transplant Congress