Extreme Phenotype Sampling and Next Generation Sequencing to Identify Genetic Variants Associated with Tacrolimus Metabolism in African American Kidney Transplant Recipients
1Hennepin Healthcare Research Institute, Minneapolis, MN, 2University of Minnesota, Minneapolis, MN, 3Nephrology, Hennepin Healthcare Research Institute, Minneapolis, MN, 4University of Nebraska Medical Center, Omaha, NE
Meeting: 2021 American Transplant Congress
Abstract number: 299
Keywords: Gene polymorphism, Genomics, Immunosuppression, Kidney transplantation
Topic: Clinical Science » Pharmacy » Non-Organ Specific: Pharmacogenomics / Pharmacokinetics
Session Information
Session Time: 10:50am-11:50am
 Presentation Time: 11:10am-11:20am
Presentation Time: 11:10am-11:20am
Location: Virtual
*Purpose: Tacrolimus (TAC) pharmacogenomics studies identified common genetic variants that alter TAC troughs suggesting strategic dosing for specific populations. Common variants and clinical factors explain ~50% of TAC trough variability in African American (AA) kidney transplant recipients. We hypothesized low-frequency variants may explain the remaining variability that may lead to improved dosing strategies for AAs.
*Methods: We repeated an Extreme Phenotype Sampling (EPS) model and expanded Next Generation Sequencing (NGS) (doi: 10.1038/s41397-018-0063-z) with substantially more samples to identify genetic variants associated with TAC troughs in AAs. AA subjects from the DeKAF Genomics and GEN03 cohorts were combined (n = 695). The study evaluated TAC troughs in the first 6 months post-transplant. EPS included 77 subjects with the highest, and 77 subjects with the lowest, dose-normalized TAC troughs after adjusting for clinical factors and CYP3A5 *3, *6, *7 variants. The NGS spanned 5.3 mB of 63 TAC related metabolism and pharmacodynamics genes, including 20 kB of flanking sequences. Variant call file (VCF) was created, SNPeff was used to assess the VCF data and Variant Effect Predictor (VEP) predicted protein functions by SIFT or Polyphen. Identified single variants were analyzed by single SNP, gene-based and subset analyses association with TAC troughs.
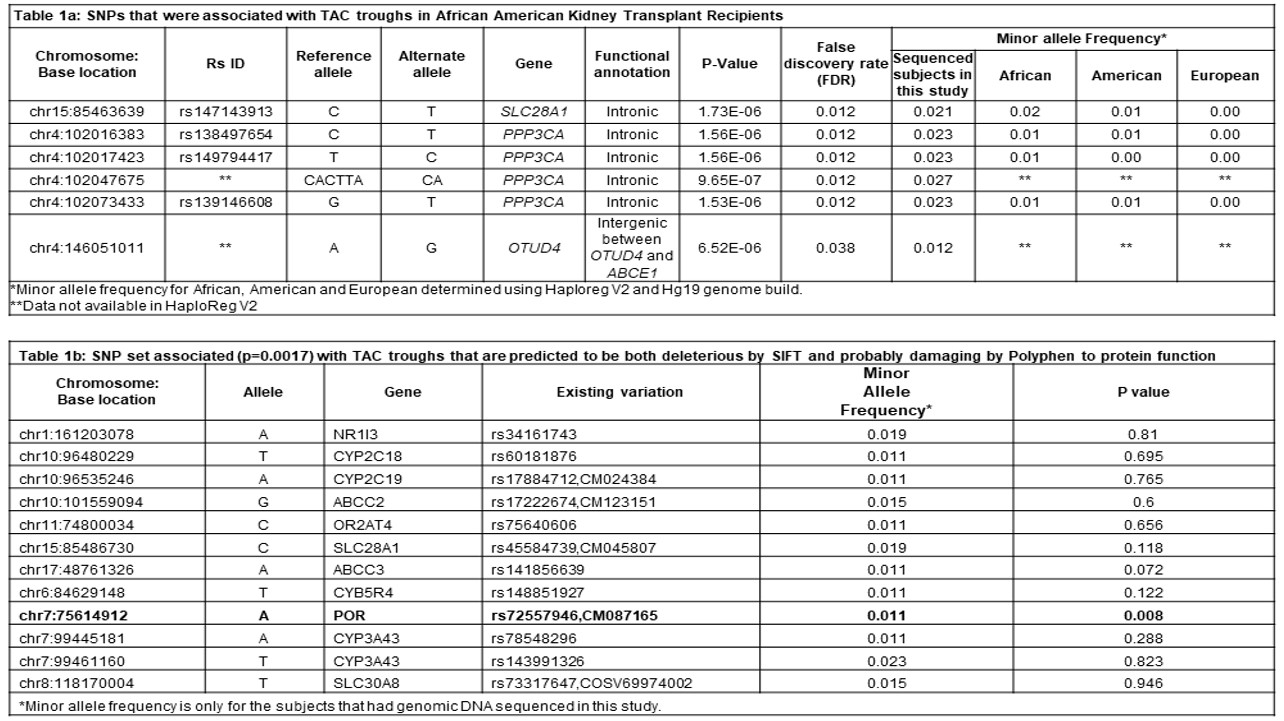
*Results: SNPeff identified 41,510 variants classified as 48% intronic, 24% intergenic, 1.3% exonic or 26.7% other. The analysis identified 6 SNPs, mostly in PPP3CA, that were associated with TAC troughs when controlling the false discovery rate at 0.05 (Table 1a). SNP set analysis of rare SNPs predicted to be deleterious by SIFT and probably damaging by Polyphen were associated with TAC troughs (p=0.0017, Table 1b). The missense variant rs72557946 in POR was also associated with TAC troughs (p=0.008).
*Conclusions: We identified novel variants associated with TAC troughs in AAs. Most of variants with TAC association were in PPP3CA which activates T-cells via NFATc and interacts with TAC. Functional analysis of these SNPs will be done in cell culture. Identification of additional SNPs which impact TAC troughs may lead to improved treatment strategies for AAs.
To cite this abstract in AMA style:
Dorr CR, Guo B, Wu B, Abrahante J, Schladt D, Remmel R, Guan W, Muthusamy A, Onyeaghala G, Pankratz N, Matas A, Mannon R, Oetting W, Jacobson P, Israni AK. Extreme Phenotype Sampling and Next Generation Sequencing to Identify Genetic Variants Associated with Tacrolimus Metabolism in African American Kidney Transplant Recipients [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/extreme-phenotype-sampling-and-next-generation-sequencing-to-identify-genetic-variants-associated-with-tacrolimus-metabolism-in-african-american-kidney-transplant-recipients/. Accessed March 9, 2026.« Back to 2021 American Transplant Congress