Molecular Diagnosis of Rejection in Liver Transplant Biopsies: The INTERLIVER Study
1Alberta Transplant Applied Genomics Centre, Edmonton, AB, Canada, 2University of Alberta, Edmonton, AB, Canada, 3., ., AB, Canada
Meeting: 2020 American Transplant Congress
Abstract number: B-130
Keywords: Gene expression, Liver transplantation, Rejection
Session Information
Session Name: Poster Session B: Liver: Immunosuppression and Rejection
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Diagnosing rejection in liver transplant biopsies is challenging given the limitations of histology e.g. low kappa values among pathologists. A Molecular Microscope® Diagnostic system (MMDx) analogous to that for kidney and heart transplants could offer improved precision and accuracy.
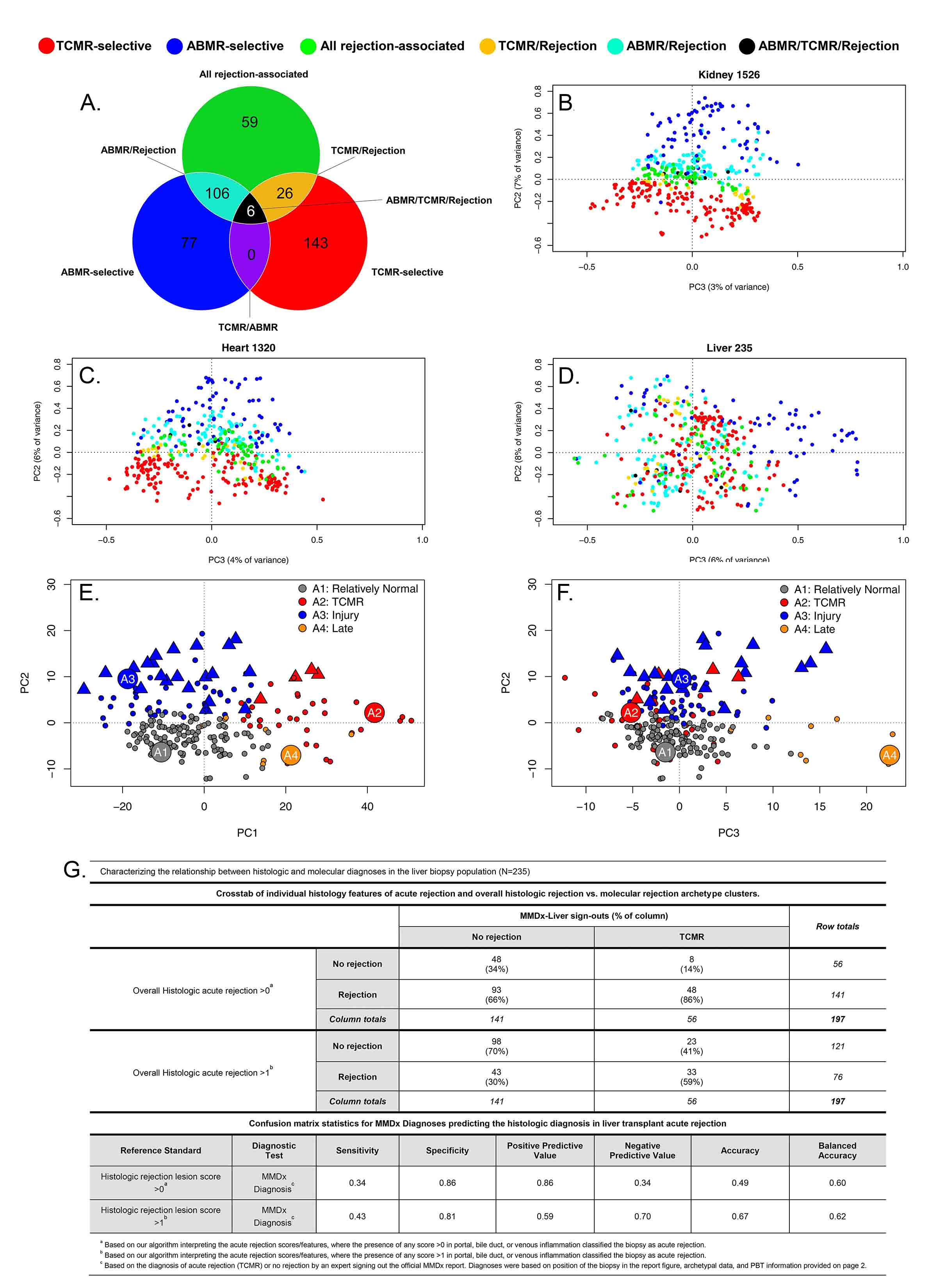
*Methods: We used gene expression microarrays to study 235 liver transplant biopsies (73% for indications) from 10 international centers (INTERLIVER ClinicalTrials.gov NCT03193151). Unsupervised archetypal analysis (AA) and principal component analysis (PCA) were used to assign molecular classes based on expression of 417 rejection-associated transcripts (RATs) derived in renal transplants (Fig1A).
*Results: Antibody-mediated rejection (ABMR) transcripts separated from T cell-mediated rejection (TCMR) transcripts in kidney and heart biopsies, but did not separate in liver (Fig1B-D), indicating that an analogous ABMR phenotype was not present or at best uncommon.
AA assigned four distinct clusters in the population: R1normal, R2TCMR, R3early injury, and R4late (Fig1E-F), differing in time post-transplant e.g. median R3injury 99 vs. R4late 3117 days. R1normal biopsies were relatively normal; R2TCMR biopsies expressed IFNG-induced TCMR-related transcripts (e.g. CXCL11); injury transcripts increased in R3injury (e.g. hypoxia inducible factor EGLN1); and R4late biopsies showed associations with atrophy-fibrosis (e.g. immunoglobulin transcripts) and injury transcripts. Groups R2-R4 were biochemically abnormal. AA scores associated with TCMR and acute injury decreased over time, while scores associated with ‘normalness’ and atrophy-fibrosis increased.
TCMR-related transcripts increased in biopsies with TCMR histology lesions (p=0.027). Biopsies called TCMR by MMDx had moderate agreement with histology (33/56 cases), but with considerable disagreement similar to that in heart transplants (R2TCMR prediction of histologic TCMR AUC=0.7) (Fig1G).
*Conclusions: By MMDx-Liver, early acute rejection (TCMR) is common in liver transplants compared to other organs, and becomes rare over time compatible with tolerogenic properties of the liver. MMDx TCMR correlates with histologic TCMR lesions, but no distinct ABMR phenotype was identified. MMDx-Liver phenotyping shows promise for improving accuracy and precision of rejection diagnoses and for guiding immunosuppressive management.
To cite this abstract in AMA style:
Madill-Thomsen KS, Halloran PF. Molecular Diagnosis of Rejection in Liver Transplant Biopsies: The INTERLIVER Study [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/molecular-diagnosis-of-rejection-in-liver-transplant-biopsies-the-interliver-study/. Accessed February 15, 2026.« Back to 2020 American Transplant Congress