Use of tCRM to Aid Diagnosis of Allograft Rejection in Kidney Transplantation
1University of California San Francisco, San Francisco, CA, 2Instituto Nacional de Ciencias Médicas y Nutrición, Mexico City, Mexico
Meeting: 2020 American Transplant Congress
Abstract number: 623
Keywords: Biopsy, Gene expression, Kidney transplantation, Rejection
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes VI
Session Type: Oral Abstract Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:45pm
 Presentation Time: 4:27pm-4:39pm
Presentation Time: 4:27pm-4:39pm
Location: Virtual
*Purpose: To apply an unbiased, quantitative, molecular profiling score for graft inflammation using standardized assays, in the context of a prospective clinical trial in kidney transplantation (KT), with a view to use this as the gold standard for acute rejection (AR) and borderline AR (bAR) diagnosis, given the wide histology-readout discrepancies between pathologists.
*Methods: We applied a tissue Common Response Module (tCRM) score in a prospective trial of 124 KT recipients, and contrasted assessment by tCRM and histology reads from 2 independent pathologists on protocol (3, 6, 12, and 24 month) and cause biopsies. 3×4 μm shaves from >500 FFPE biopsies were processed for qPCR (Fluidigm) for 11 genes. The tCRM score was calculated for each biopsy specimen using the geometric means of gene expression of these 11 genes (>2.2 bAR; >4 AR). Results were correlated with graft function, DSA, and pathology.
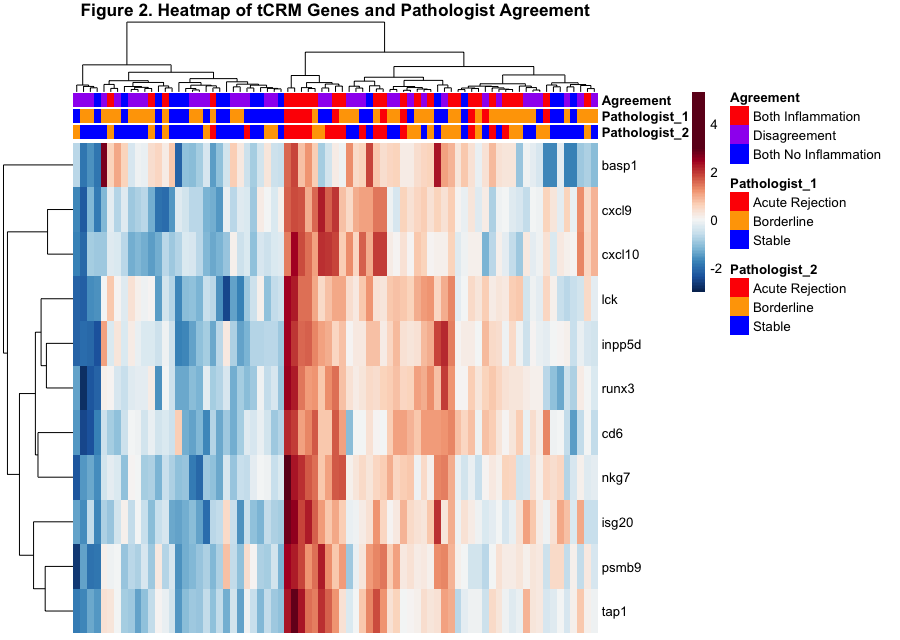
*Results: The tCRM score was significantly elevated in histological categories that aligned between both pathologists diagnosing AR (3.7 ± 2.8) vs. no AR (1.5 ± 1.0) (Figure 1). Unsupervised clustering of tCRM gene expression across biopsies where pathologists had discrepant scores is shown in Figure 2, and ~45% of pathology reads had discordance, with maximal discordance between cross-pathologist reads of bAR vs. stable. Maximal histology agreement was noted for AR, with high correlation with molecular AR by tCRM. Conversely, concordant stable samples by histology had statistically significant low tCRM scores.
*Conclusions: Accurate, quantitative, and unbiased assessment of rejection on the clinical biopsy sample is critical to evaluate outcomes in a clinical trial. Maximal discrepancy is noted between pathologists for bAR vs. stable reads, and the unbiased molecular readout of rejection by tCRM provides a critical tie-breaker for final clinical diagnosis. The tCRM assay can be assessed by Fluidigm or Nanostring, only uses 11 genes, and utilizes a shave off the existing FFPE block. We propose that the tCRM quantitative score should be utilized for routine pathology assessment, as an unbiased assay to support clinical decision making.
To cite this abstract in AMA style:
Zarinsefat A, Liberto J, Damm I, Ramirez C, Guerra J, Alberu J, Sarwal M. Use of tCRM to Aid Diagnosis of Allograft Rejection in Kidney Transplantation [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/use-of-tcrm-to-aid-diagnosis-of-allograft-rejection-in-kidney-transplantation/. Accessed February 19, 2026.« Back to 2020 American Transplant Congress