microPOTS: A New Single Cell Proteomic Method for Profiling Human Kidney Transplant Biopsy Tissue
1University of California San Francisco, San Francisco, CA, 2Pacific Northwest National Laboratory, Richland, WA
Meeting: 2020 American Transplant Congress
Abstract number: 214
Keywords: Biopsy, Kidney, Methodology, Peptides
Session Information
Session Name: Biomarker Discovery and Immune Modulation
Session Type: Oral Abstract Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:45pm
 Presentation Time: 3:27pm-3:39pm
Presentation Time: 3:27pm-3:39pm
Location: Virtual
*Purpose: The purpose of this study was to interrogate acute rejection (AR) in the renal allograft at single cell level. Despite advances in single cell technology at the RNA level, proteomics assays cannot reduce assay dimensionality robustly below 500 cells. Our aim was to develop a novel protocol for robust single cell proteomics (scProteomics) to enable interrogation of as few as 1-10 cells/kidney biopsy compartment, from very small amount of tissue, as available from half of an 18G kidney transplant biopsy core.
*Methods: Technological advances have been done with the development of a new microPOTS technique, with custom laser capture microdissection glomerular (G) and proximal tubular (PT) cells from 10 kidney transplant biopsy cores collected in optimal cutting temperature (OCT) medium. Liquid-chromatography mass-spectrometry was optimized in microPOTS for analysis of ultra-low amounts of protein extracted from each of the micro-dissected compartments. Raw data was analyzed with MaxQuant, compartment specific enrichment analysis was performed with matched bulk proteomic and single-cell RNASeq data from the same kidney samples.
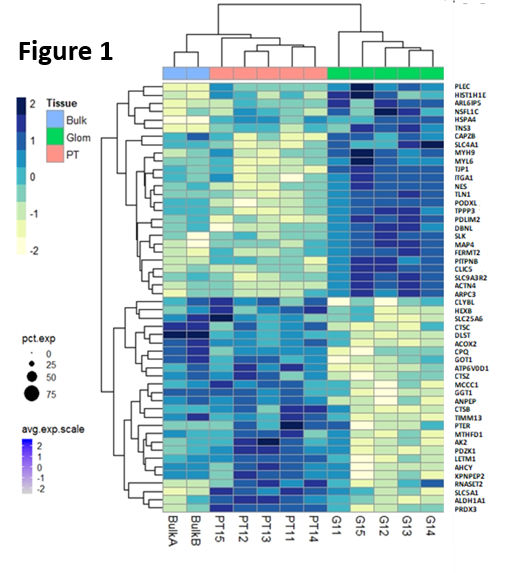
*Results: Ten micron thick OCT sections of kidney were optimized for laser capture, and resulted in <30 cells/ compartment and ~2560 proteins/sample. Replicates were highly reproducible; (r) =0.93, P<0.0001. microPOTS resulted in unique compartment specific proteins, missing in the paired bulk proteomic analysis. 208 proteins were highly enriched in the glomerulus, with known podocyte markers present (PODXL, CLIC5) with 67 proteins being unique to the glomerulus. 247 proteins were enriched (>2 fold) in the proximal tubule, with known tubule markers (PDZK1, ANPEP) and 25 proteins were unique ( most enriched proteins for each compartment shown in Fig.1). Some unique compartment specific proteins have not been reported to date in the literature for these kidney regions. IHC stains are underway to confirm the sub-compartment localization these novel proteins.
*Conclusions: The first near single-cell proteomic protocol has been developed and optimized for use on human kidney tissue samples, providing a new spatial technology to understand cell and compartment specific differences in rejection. Unique glomerular and proximal tubular proteins have been identified which are currently missed by scRNASeq and bulk proteomics, and these appear to have low abundance, but may have high biological relevance in the pathogenesis of disease and rejection.
To cite this abstract in AMA style:
Sigdel T, Piehowski P, Roy S, Liberto J, Schroeder A, Rashmi P, Sur S, Damm I, Qian W, Sarwal MM. microPOTS: A New Single Cell Proteomic Method for Profiling Human Kidney Transplant Biopsy Tissue [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/micropots-a-new-single-cell-proteomic-method-for-profiling-human-kidney-transplant-biopsy-tissue/. Accessed February 28, 2026.« Back to 2020 American Transplant Congress