Molecular Classifiers Can Outperform the Flawed Histologic “Gold Standard” on Which They Are Trained
University of Alberta, Edmonton, Canada.
Meeting: 2018 American Transplant Congress
Abstract number: A56
Keywords: Gene expression, Kidney transplantation
Session Information
Session Name: Poster Session A: Biomarkers, Immune Monitoring and Outcomes
Session Type: Poster Session
Date: Saturday, June 2, 2018
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall 4EF
The histologic diagnosis of rejection in organ transplants is known to be an imperfect gold standard. Therefore, it might be assumed that molecular classifiers built using such diagnoses could, at best, be no better than the training set diagnoses. If true, the only utility of a molecular test would be in situations where it was cheaper or less invasive than the biopsy, e.g. as a blood screening test to see if a biopsy was necessary. In our "Molecular Microscope" system, we have taken a different approach, suggesting that machine learning-based molecular models, though trained on flawed labels, make more accurate diagnoses than histology in both the original training set and in future biopsies. Here, we present results from simulation studies showing how this is possible.
From a data set of 1208 kidney biopsies, we chose 60 that were almost certainly antibody-mediated rejection (ABMR) (they had histologic ABMR and high molecular scores for the ABMR-related gene ROBO4), and 60 that were almost certainly non-rejecting (histologically clean with low ROBO4 scores). This was to establish a set of biopsies in which the truth was known with at least a fair degree of certainty.
Given the absence of diagnostically intermediate biopsies, a classifier built using these samples can predict the correct diagnosis with 100% accuracy (Fig 1A – all results are from test sets using 10-fold cross-validation). The simulation added increasing levels of error in the training set diagnostic labels (Fig 1B-F), and measured classifier accuracy (test set error) in comparison to the "truth", i.e. the diagnoses before the labels were altered. Even with 20% of diagnoses mislabeled in the training sets (Fig 1C), the classifier is able to make the correct diagnosis with 100% accuracy. As the accuracy of the gold standard decreased, the classifier became increasingly uncertain in its diagnoses, until at some point the distributions of ABMRs and non-ABMRs start to overlap (Fig 1D-F).
In summary, strong molecular signals associated with rejection phenotypes, combined with diagnostic labels that are at least "mostly" correct, allow for the generation of machine learning models that are more accurate than histology alone.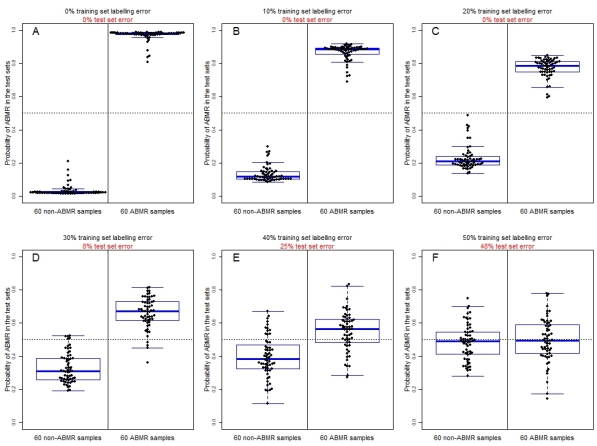
CITATION INFORMATION: Reeve J., Halloran P. Molecular Classifiers Can Outperform the Flawed Histologic “Gold Standard” on Which They Are Trained Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Reeve J, Halloran P. Molecular Classifiers Can Outperform the Flawed Histologic “Gold Standard” on Which They Are Trained [abstract]. https://atcmeetingabstracts.com/abstract/molecular-classifiers-can-outperform-the-flawed-histologic-gold-standard-on-which-they-are-trained/. Accessed February 5, 2026.« Back to 2018 American Transplant Congress
