Quantitative Gene Expression Analysis of CRM-Panel Genes Using FFPE Kidney Tissues for Transplant Injuries.
University of California San Francisco, San Francisco
Meeting: 2017 American Transplant Congress
Abstract number: A131
Keywords: Biopsy, Gene expression, Rejection
Session Information
Session Name: Poster Session A: Diagnostics/Biomarkers Session I
Session Type: Poster Session
Date: Saturday, April 29, 2017
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall D1
Background: A common immune response module (CRM) of 11 genes was identified to define acute rejection (AR) through meta-analyses of 1030 different engrafted tissues including kidney, heart, lung and liver. A CRM score for graft injury detection was defined for standard QPCR assays using samples submerged in RNA stabilizing solution. In this study, we validated the utility of CRM-panel of genes as a metric for sub-clinical and clinical transplant injury in FFPE kidney tx tissues using an assay that uses barcoded oligos for mRNA transcript quantification.
Method: Carefully selected FFPE tissues at the time of graft dysfunction event from 40 kidney tx patients (8 in each of normal functioning (NL), antibody mediated rejection (AMR), T cell mediated rejection (TCMR), borderline changes (BL), and BK virus nephropathy (BKVN). The RNA extraction from FFPE sections was optimized. Barcoded Codesets for each of 11 CRM gene-set and a set of reference genes (GAPDH, GUSB, HPRT1, LDHA, and TBP) used in the study were designed for gene expression analysis using nCounter system. Data normalization was performed using positive controls and housekeeping genes.
Results: Consistent with prior fresh tissues studies and conventional qPCR, the CRM score as quantified by barcoded oligos from FFPE kidney allograft tissue was able to differentiate various types of allograft injury from stable function. The average CRM score was 1.5, 8.3, 4.4, 5.6, and 4.4 for NL, TCMR, AMR, BL and BKVN, respectively. When compared to NL, the CRM score was significantly greater is TCMR (p = 0.0080), AMR (p = 0.0079), and BKVN (p = 0.0084) and B (p = 0.17).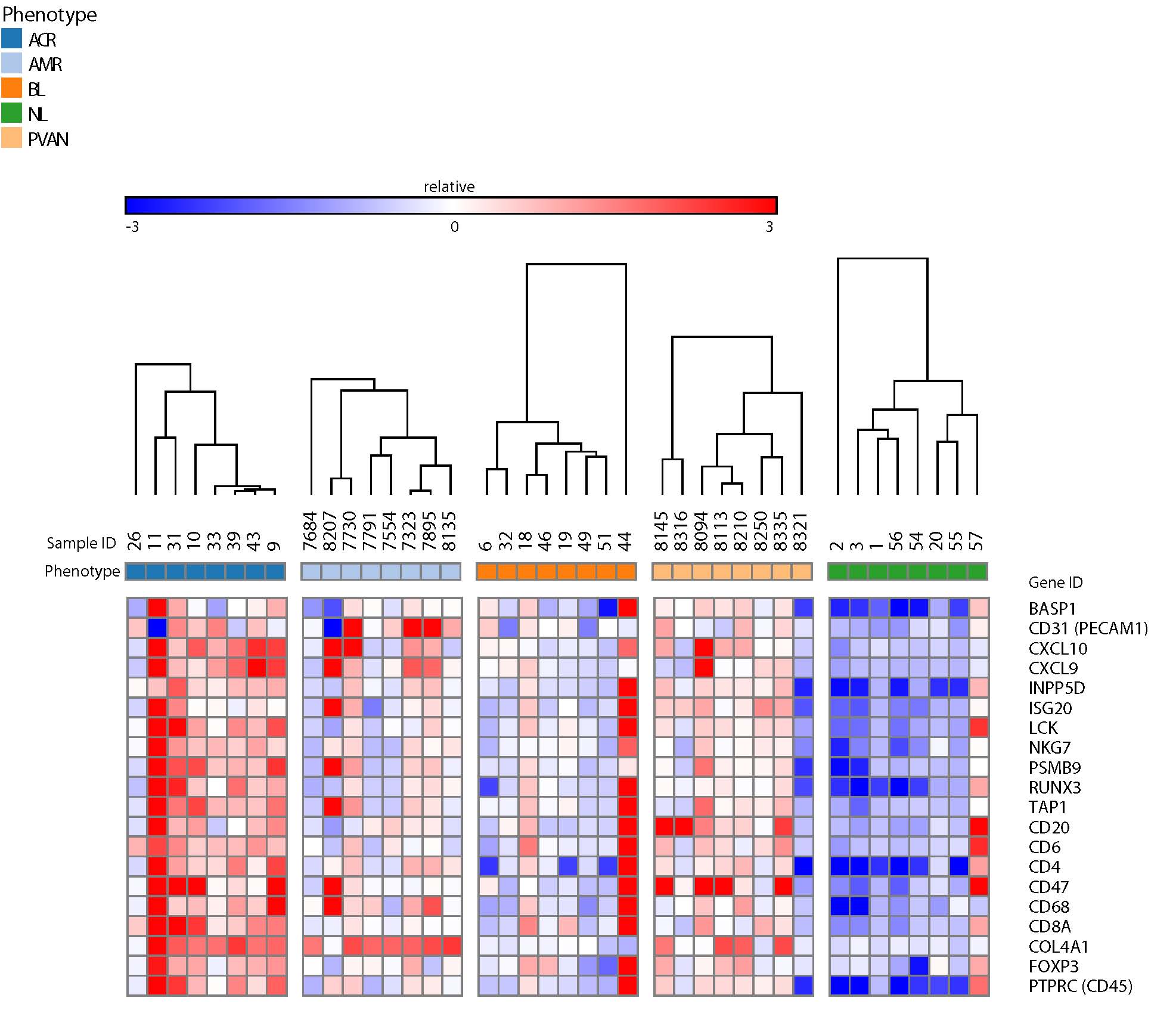 Conclusion: This study successfully demonstrates an extension utility of the tissue CRM gene assay for quantifying tissue immune burden in tx kidney biopsies, and can be used to define absolute molecular inflammation on protocol and for cause biopsies in a rapid and cost effective approach.
Conclusion: This study successfully demonstrates an extension utility of the tissue CRM gene assay for quantifying tissue immune burden in tx kidney biopsies, and can be used to define absolute molecular inflammation on protocol and for cause biopsies in a rapid and cost effective approach.
CITATION INFORMATION: Sigdel T, Nguyen M, Dobi D, Hsieh S.-C, Liberto J, Vincenti F, Sarwal M, Laszik Z. Quantitative Gene Expression Analysis of CRM-Panel Genes Using FFPE Kidney Tissues for Transplant Injuries. Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Sigdel T, Nguyen M, Dobi D, Hsieh S-C, Liberto J, Vincenti F, Sarwal M, Laszik Z. Quantitative Gene Expression Analysis of CRM-Panel Genes Using FFPE Kidney Tissues for Transplant Injuries. [abstract]. Am J Transplant. 2017; 17 (suppl 3). https://atcmeetingabstracts.com/abstract/quantitative-gene-expression-analysis-of-crm-panel-genes-using-ffpe-kidney-tissues-for-transplant-injuries/. Accessed February 11, 2026.« Back to 2017 American Transplant Congress
