Microarray Analysis of Rejection in Formalin-Fixed Paraffin-Embedded Biopsies: Similarities and Disparities with Conventionally Stabilized Biopsies.
Alberta Transplant Applied Genomics Centre, University of Alberta, Edmonton, AB, Canada
Meeting: 2017 American Transplant Congress
Abstract number: A22
Keywords: Gene expression, Kidney transplantation, Methodology, Rejection
Session Information
Session Name: Poster Session A: Antibody Mediated Rejection in Kidney Transplant Recipients I
Session Type: Poster Session
Date: Saturday, April 29, 2017
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall D1
Despite potential for performing molecular diagnostics on formalin-fixed paraffin embedded (FFPE) biopsies, there has been no systematic comparison of microarray gene expression data obtained from FFPE versus immediately stabilized (RNAlater) kidney transplant biopsies.
We selected 70 FFPE samples (from 68 patients) representing T cell-mediated, antibody-mediated rejection (TCMR, ABMR) and no rejection, and compared them with RNAlater samples obtained from the same biopsy at the same time. Microarray and RNA performance was evaluated using rejection-associated transcripts (RATs) – a union of the top ABMR, TCMR and all rejection algorithms obtained from each method, and applied them in Principal Component Analysis. The average yield of total RNA was 0.46 ug from FFPE and 4.1ug from RNAlater samples, i.e. FFPE gave ~9 times less RNA. Samples were run on PrimeView chips (Affymetrix). Because of different labeled products, the FFPE and RNAlater samples were analyzed separately before comparisons.
Figure 1 shows the distribution of 70 FFPE and RNAlater samples analyzed for expression of 577 RNAlater and 551 FFPE RATs. 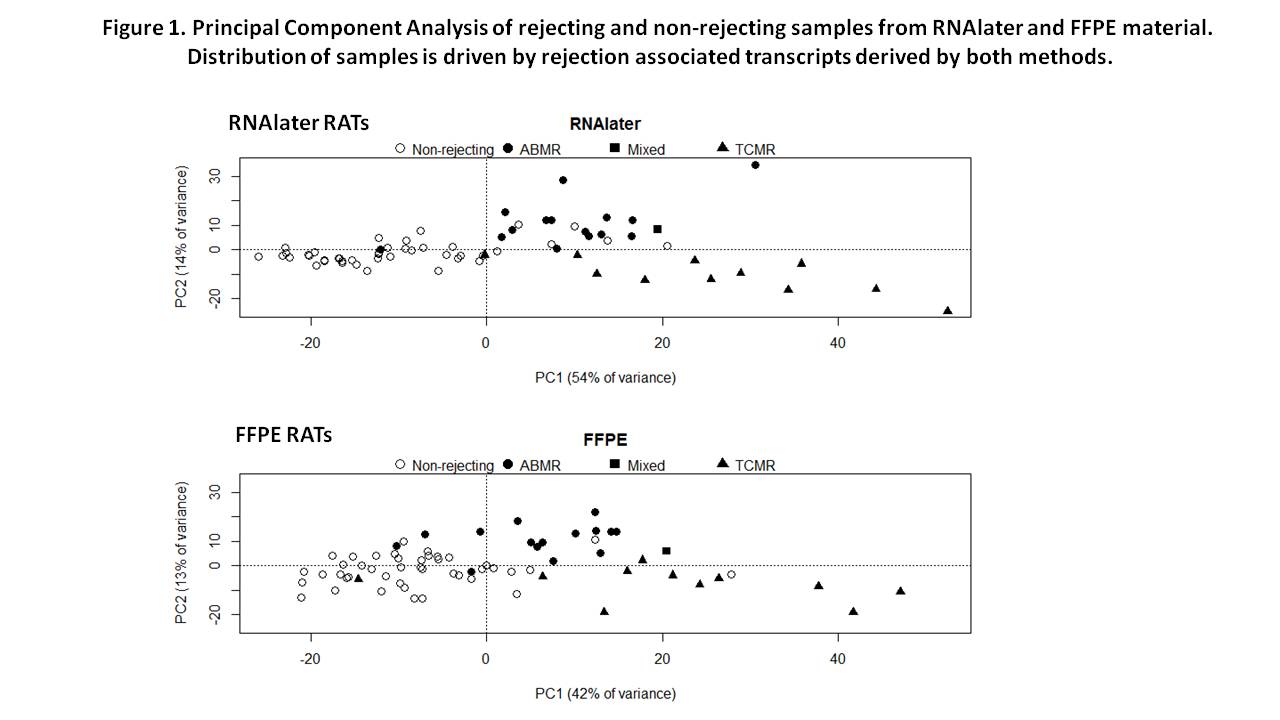 The distribution of rejection phenotypes was similar in the two sets of samples. However, the FFPE results were noisier, particularly for relatively normal biopsies, and gave some false positive associations. The association strength of RNAlater RATs with rejection phenotypes was much better than with FFPE RATs, probably reflecting RNA damage in the FFPE samples. RNAlater yielded 10 times as many significant transcripts in comparison of all rejection vs. no rejection. Moreover, only 25% of RATs were shared by RNAlater and FFPE methods.
The distribution of rejection phenotypes was similar in the two sets of samples. However, the FFPE results were noisier, particularly for relatively normal biopsies, and gave some false positive associations. The association strength of RNAlater RATs with rejection phenotypes was much better than with FFPE RATs, probably reflecting RNA damage in the FFPE samples. RNAlater yielded 10 times as many significant transcripts in comparison of all rejection vs. no rejection. Moreover, only 25% of RATs were shared by RNAlater and FFPE methods.
Conclusion: The method of choice for molecular studies should be intact RNA from biopsies stabilized in RNAlater or snap frozen, with FFPE as a second choice. FFPE damage to RNA creates some potential false positive and false negative artifacts. The advantage of FFPE samples – use of the sample remaining after histology processing – must be weighed against the disadvantages of smaller amounts of damaged RNA, with potential increase in sampling error.
CITATION INFORMATION: Famulski K, Revee J, Halloran P. Microarray Analysis of Rejection in Formalin-Fixed Paraffin-Embedded Biopsies: Similarities and Disparities with Conventionally Stabilized Biopsies. Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Famulski K, Revee J, Halloran P. Microarray Analysis of Rejection in Formalin-Fixed Paraffin-Embedded Biopsies: Similarities and Disparities with Conventionally Stabilized Biopsies. [abstract]. Am J Transplant. 2017; 17 (suppl 3). https://atcmeetingabstracts.com/abstract/microarray-analysis-of-rejection-in-formalin-fixed-paraffin-embedded-biopsies-similarities-and-disparities-with-conventionally-stabilized-biopsies/. Accessed March 3, 2026.« Back to 2017 American Transplant Congress
