Proteoforms: Novel Biomarkers for Acute Rejection in Liver Transplant Recipients.
1Northwestern, Chicago
2UPMC, Pittsburgh.
Meeting: 2016 American Transplant Congress
Abstract number: 141
Keywords: Gene expression, Liver transplantation, Rejection
Session Information
Session Name: Concurrent Session: Liver: Immunosuppression and Rejection
Session Type: Concurrent Session
Date: Sunday, June 12, 2016
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:54pm-5:06pm
Presentation Time: 4:54pm-5:06pm
Location: Room 312
Background: Biomarker profiles diagnostic of acute rejection (AR) in liver transplant (LT) recipients could enhance the diagnosis and management of recipients. Our aim was to identify diagnostic protein signatures for AR in PBMC, using a novel top down proteomics methodology. In this approach, proteins are kept intact prior to analysis by an integrated separations and mass spectrometric platform, allowing evaluation of protein primary structure and post-translational modifications (proteoforms).
Methods:PBMC lysates from non-HCV+ LT recipients undergoing liver biopsy were processed by a technique similar to SDS-PAGE (GELFrEE) to create a single fraction containing all proteins from 0-30 kDa with high recovery. After SDS removal, this protein fraction was analyzed by top-down proteomics in 3 steps: 1) Nanocapillary liquid chromatography coupled with a Orbitrap Elite mass spectrometer, 2) processing data using ProSight software to identify proteoforms by database searching, and 3) processing data through a linear hierarchical model matching the study design to quantify proteoform fold changes in AR vs. Tx (normal liver function) vs. ADNR (acute dysfunction without rejection).
Results: 24 patients, age 60±15.6 years, 66.6% male, mean 3.7±3.3 year from LT. Differentially expressed proteoforms were seen in patients with AR vs. TX and AR vs. ADNR 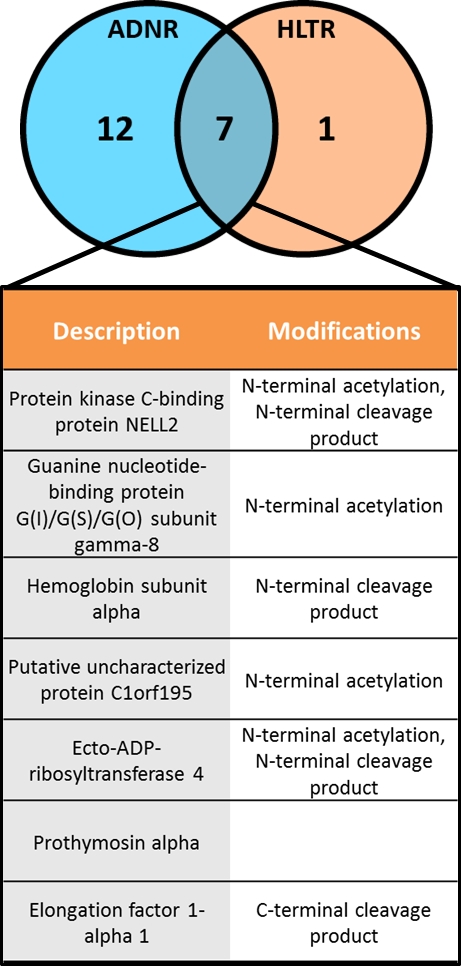 . As shown, seven proteins with distinct post-translational modifications were increased in AR compared to both TX and ADNR, suggesting a distinct proteoform signature of AR in LT recipients. Mapping analysis of the seven proteins back to genes through PANTHER (Protein Analysis Through Evolutionary Relationships) revealed multiple signaling pathways, including inflammation mediated by cytokines and chemokines.
. As shown, seven proteins with distinct post-translational modifications were increased in AR compared to both TX and ADNR, suggesting a distinct proteoform signature of AR in LT recipients. Mapping analysis of the seven proteins back to genes through PANTHER (Protein Analysis Through Evolutionary Relationships) revealed multiple signaling pathways, including inflammation mediated by cytokines and chemokines.
Conclusion: We have performed a unique methodology (top down proteomics) revealing a proteoform signature for AR distinct from other causes of graft dysfunction. Larger studies are needed to validate these signatures and test their predictive value for use in clinical management.
CITATION INFORMATION: Levitsky J, Toby T, Kim K, Ntai I, Demetris A, Abecassis M, Thomas P, Kelleher N. Proteoforms: Novel Biomarkers for Acute Rejection in Liver Transplant Recipients. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Levitsky J, Toby T, Kim K, Ntai I, Demetris A, Abecassis M, Thomas P, Kelleher N. Proteoforms: Novel Biomarkers for Acute Rejection in Liver Transplant Recipients. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/proteoforms-novel-biomarkers-for-acute-rejection-in-liver-transplant-recipients/. Accessed February 24, 2026.« Back to 2016 American Transplant Congress
