Using Ensembles of Machine Learning Classifiers to Maximize the Accuracy and Stability of Molecular Biopsy Interpretation
1University of Alberta, Edmonton, AB, Canada, 2Alberta Transplant Applied Genomics Centre, University of Alberta, Edmonton AB, AB, Canada, 3University of Alberta, Edmonton AB, AB, Canada
Meeting: 2019 American Transplant Congress
Abstract number: 318
Keywords: Gene expression, Kidney, Kidney transplantation, Rejection
Session Information
Session Name: Concurrent Session: Kidney Chronic Antibody Mediated Rejection
Session Type: Concurrent Session
Date: Monday, June 3, 2019
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:54pm-5:06pm
Presentation Time: 4:54pm-5:06pm
Location: Ballroom B
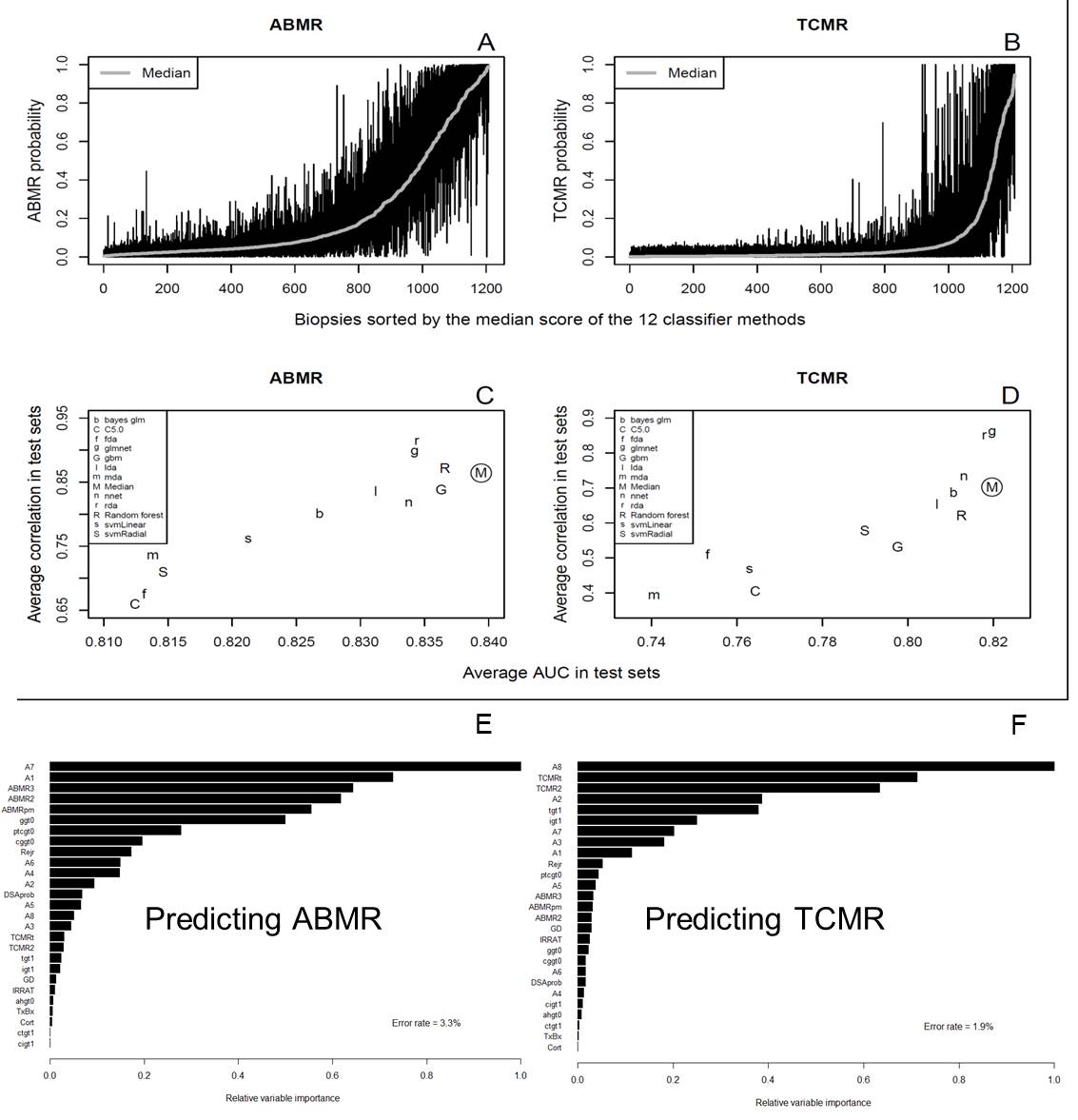
*Purpose: The Molecular Microscope diagnostic system (MMDx), based on microarray gene expression, uses ensembles of machine learning classifiers rather than single genes, gene sets, or classifiers, to maximize the accuracy of rejection diagnoses and injury assessment. We tested its accuracy and stability, and developed an automated system for generating molecular reports on kidney transplant biopsies.
*Methods: We evaluated the ensembles’ accuracy (agreement with histology) and stability (correlation of predictions based on multiple training sets). 1679 kidney transplant biopsies were repeatedly split at random into two training sets (N=600 each) and a test set (N=479). Classifiers were developed in each training set, and predictions for ABMR and TCMR made in the test sets. Twelve separate machine learning methods and their median were evaluated. In a separate analysis, a random forest classifier was used to predict the report sign-outs of an expert clinician.
*Results: There was considerable variation between the 12 classifier methods for any given biopsy (Figure 1A and B). The median had a higher accuracy than any of the individual classifiers, and was among the most stable (highest correlation between predictions from separate random training sets – Figure 1C and D). A random forest classifier was used to predict the sign-out of an expert evaluator (Figure 1E and F – abbreviations are explained on the MMDx report). Accuracies for the expert’s molecular TCMR and ABMR diagnoses were ~98 and 97% respectively. Most disagreements were in biopsies near diagnostic thresholds.
Considerable disagreement with histology persists, which is expected given the noise in histology assessments. The balanced accuracy of MMDx signouts for pathology diagnoses of TCMR and ABMR was about 75%.
*Conclusions: In our data set, ensembles of machine learning classifiers generate diagnoses that are both more accurate than the best individual classifiers, and nearly as stable as the best. This result is expected from the machine learning literature, since different methods will tend to make different types mistakes, and taking the medians cancels out the worst estimates. Similar ensembles (random forests) can be used to create automated report sign-outs that agree with an expert observer 97-98% of the time. Disagreement with histology will persist, largely due to the known noise in histology assessments (ClinicalTrials.gov NCT01299168).
To cite this abstract in AMA style:
Reeve J, Madill-Thomsen KS. Using Ensembles of Machine Learning Classifiers to Maximize the Accuracy and Stability of Molecular Biopsy Interpretation [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/using-ensembles-of-machine-learning-classifiers-to-maximize-the-accuracy-and-stability-of-molecular-biopsy-interpretation/. Accessed February 19, 2026.« Back to 2019 American Transplant Congress