Unifying Interpretation of Molecular HLA Typing Data between the Solid Organ Allocation Match Run and Virtual Crossmatch
1Tulane University School of Medicine, New Orleans, LA, 2Cedars-Sinai Heart Institute, Los Angeles, CA, 3Baylor Scott & White, Temple, TX
Meeting: 2019 American Transplant Congress
Abstract number: A214
Keywords: Alloantibodies, Allocation, Histocompatibility, Kidney transplantation
Session Information
Session Name: Poster Session A: Kidney Deceased Donor Allocation
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: Virtual crossmatch compares a transplant candidate’s HLA antibody assay results to the HLA typing of the donor before an organ offer is accepted. While HLA typing is now performed by molecular methods in histocompatibility laboratories, the United Network for Organ Sharing (UNOS) UNet system that generates the match run to determine organ offers allows for entry of only two donor HLA antigens per locus and does not capture precise information about molecular HLA typing ambiguity.
*Methods: We have developed a web application to compute virtual crossmatch and the histocompatibility component of match run using molecular HLA typing data in IPD-IMGT/HLA nomenclature, using genotype lists or multiple allele codes to represent ambiguity. The application computes the probability of each potential conflicting HLA specificity, both at two-field allele and antigen level, based on high resolution allele frequencies from 26 US populations obtained from the National Marrow Donor Program.
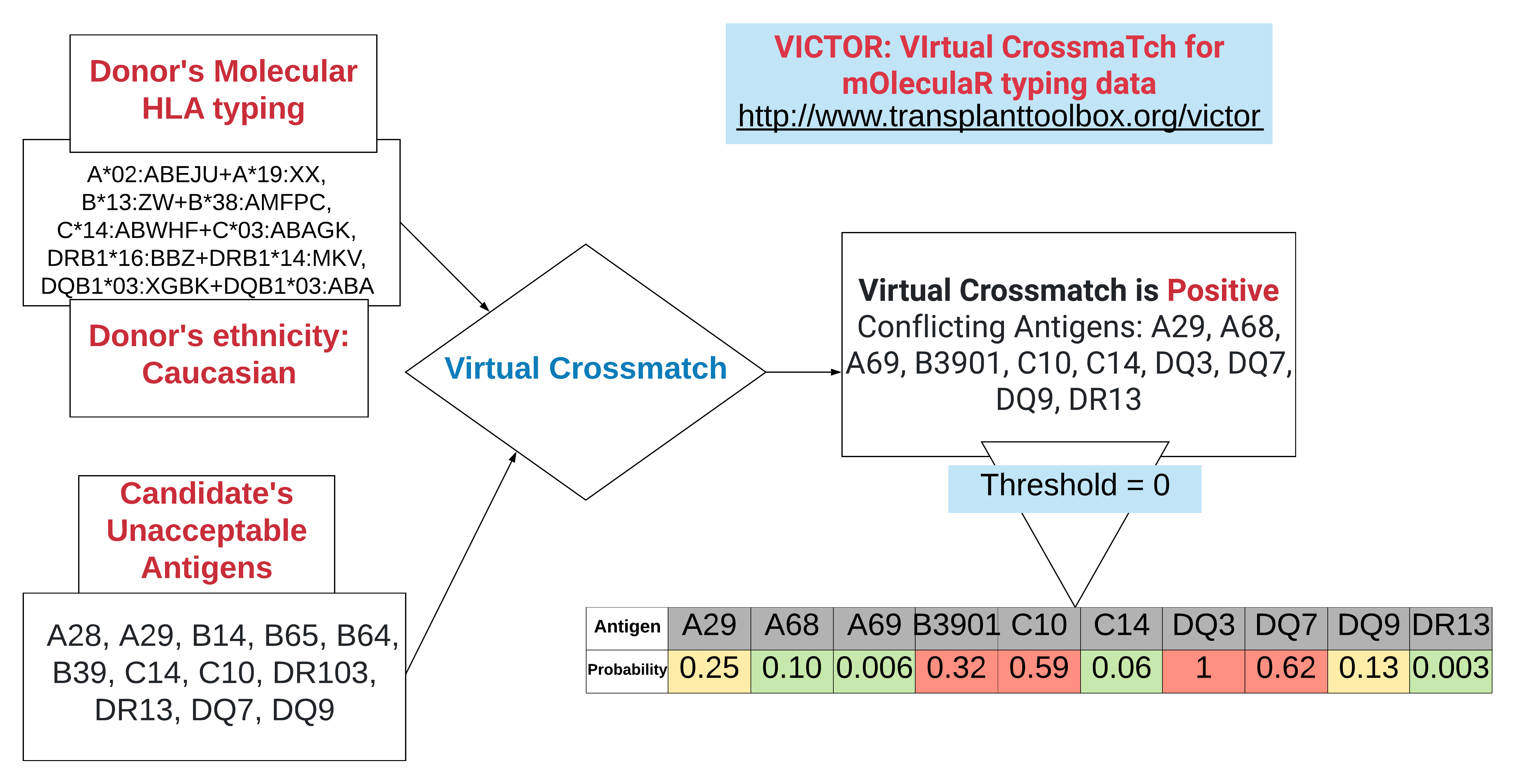
*Results: Our analysis of simulated donor-candidate compatibility assessments reveals the risks of conflicts for alternative less probable HLA antigens and alleles of the donor that cannot be listed in the UNet system. We detail cases where organ offers from the match run would be turned down upon closer examination of the molecular HLA typing data in virtual crossmatch. In cases where virtual crossmatch fails to assess that unacceptable antigens are likely to present, organs may be shipped to incompatible recipients. We show how analysis of molecular HLA typing can be aided by a computational decision support system. The VIrtual CrossmaTch for mOleculaR HLA typing (VICTOR) application can be accessed at http://www.transplanttoolbox.org/victor. Algorithm for computation of virtual crossmatch with risk stratification by VICTOR for ambiguous HLA typing is shown in the figure below.
*Conclusions: HLA typing ambiguity has been an unappreciated source of immunological risk in transplantation. This application automates translation of HLA data between antigen-based and molecular nomenclature systems to streamline virtual crossmatch in the short time available to accept organ offers. A standardized interpretation of molecular HLA typing data could be integrated in the match run along with epitope-based interpretations to conduct more comprehensive risk assessments.
To cite this abstract in AMA style:
Kaur N, Kransdorf E, Pando M, Gragert L. Unifying Interpretation of Molecular HLA Typing Data between the Solid Organ Allocation Match Run and Virtual Crossmatch [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/unifying-interpretation-of-molecular-hla-typing-data-between-the-solid-organ-allocation-match-run-and-virtual-crossmatch/. Accessed March 3, 2026.« Back to 2019 American Transplant Congress