The Molecular Landscape of Bk Polyomavirus Infection in Kidney Transplant Recipients at Single Cell Resolution
University of Michigan, Ann Arbor, MI
Meeting: 2022 American Transplant Congress
Abstract number: 252
Keywords: Genomics
Topic: Clinical Science » Infection Disease » 26 - Kidney: Polyoma
Session Information
Session Name: Kidney: Polyoma Infections
Session Type: Rapid Fire Oral Abstract
Date: Monday, June 6, 2022
Session Time: 3:30pm-5:00pm
 Presentation Time: 3:30pm-3:40pm
Presentation Time: 3:30pm-3:40pm
Location: Hynes Room 309
*Purpose: BK polyomavirus has devastating consequences in patients with kidney transplants. While in vitro studies in primary epithelial cell lines, and bulk transcriptomic studies in human kidney biopsies of patients with BKVAN have improved our understanding of the virus, they remain limited due to the inherent limitation of such technologies.
*Methods: Single cell-specific transcriptional profiles from kidney biopsies of fourteen patients with BK viremia in the first year were compared with fourteen first-year healthy transplant controls. Three biopsies were collected when the trajectory of BK viremia was ‘peaking,’ and eleven biopsies were collected during the ‘resolving’ phase of the infection after net immunosuppression reduction.
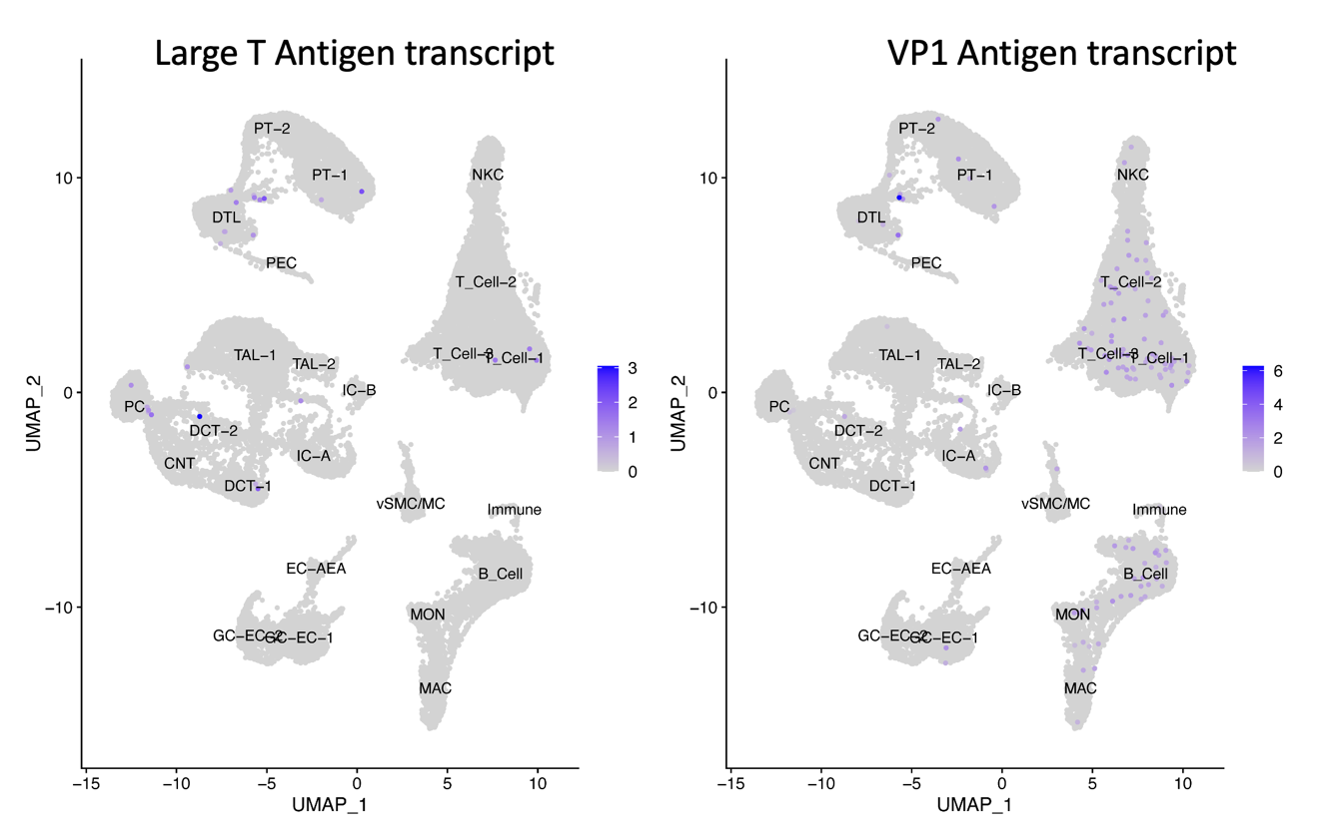
*Results: Results: Viral transcripts associated with productive infection, i.e., LT1 and VP1, were detected in kidneys from two of three patients with peaking infection and confirmed by SV40 immunohistochemical staining. Those viral transcripts were detected in various renal cell types confirmed by immunohistochemical studies. Transcripts were also detected in immune cells, notably T and B cells. LT1 mRNA expression was associated with upregulation of genes related to pro-survival, endocytosis, cell-cell adhesion, and increased metabolic processes pathways. At the same time there was downregulation of Class 1 and Class 2 HLA molecules. Among renal cells with VP1 transcript expression, was an enrichment of genes associated with an increase in TGF-b/SMAD signaling, but downregulation of genes associated with tissues specific immune responses and other metabolic processes. In tubular cells, there was decreased expression of EGF and UMOD transcripts during ‘peaking’ (vs. resolving infection) and increased expression of HLA class 2 molecules consistent with tubular injury and increased immunogenicity, respectively. Analysis of the BK virus non-coding control (NCCR) region suggested the presence of both archetype and rearranged variants of the virus in both patients with BK nephropathy and occasionally in the same infected cell.
*Conclusions: Our single-cell transcriptional analysis offers novel mechanistic insights into BK polyomavirus infection, including identifying transcriptional machinery of kidney-specific and immune cells in peaking and resolving polyomavirus infection. Furthermore, the presence of viral transcripts in immune cells and a critical analysis of the NCCR region offers potential mechanistic insight into persistent BK polyomavirus infection.
To cite this abstract in AMA style:
McCown PJ, Farkash E, Alakwaa F, Eichinger F, Menon R, Otto E, Berthier CC, Woodside KJ, Norman S, Kretzler M, Naik A. The Molecular Landscape of Bk Polyomavirus Infection in Kidney Transplant Recipients at Single Cell Resolution [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/the-molecular-landscape-of-bk-polyomavirus-infection-in-kidney-transplant-recipients-at-single-cell-resolution/. Accessed March 7, 2026.« Back to 2022 American Transplant Congress