Single Nuclei RNA-Sequencing Identifies Distinct Immune Cell Profile in Kidney Allograft Fibrosis
1University of Maryland Baltimore, Baltimore, MD, 2UTHSC, Memphis, TN, 3Weill Cornell Medicine, New York, NY
Meeting: 2022 American Transplant Congress
Abstract number: 679
Keywords: Fibrosis, Kidney, Kidney transplantation
Topic: Basic Science » Basic Clinical Science » 19 - Chronic Organ Rejection
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:30pm-7:00pm
Presentation Time: 5:30pm-7:00pm
Location: Hynes Halls C & D
*Purpose: A major obstacle after kidney transplantation (KT) is chronic allograft dysfunction characterized by interstitial fibrosis and tubular atrophy (IFTA). Despite extensive research, the cellular immune response associated with IFTA is unclear. Single nuclei RNA sequencing (snRNA-seq) is a powerful tool to uncover the immune landscape.
*Methods: snRNA-seq was performed for normal (N=3) and IFTA (N=5) human allografts characterized by Banff classifications (all were functional at ≥15mos posttransplant). Sequences were aligned using CellRanger and integrated using the R package Seurat. Ligand receptor analysis utilized the LRdb and an interaction score was computed using machine learning. XY chromosome linked gene expression analysis was used to determine cellular origins using sex-mismatched KTs.
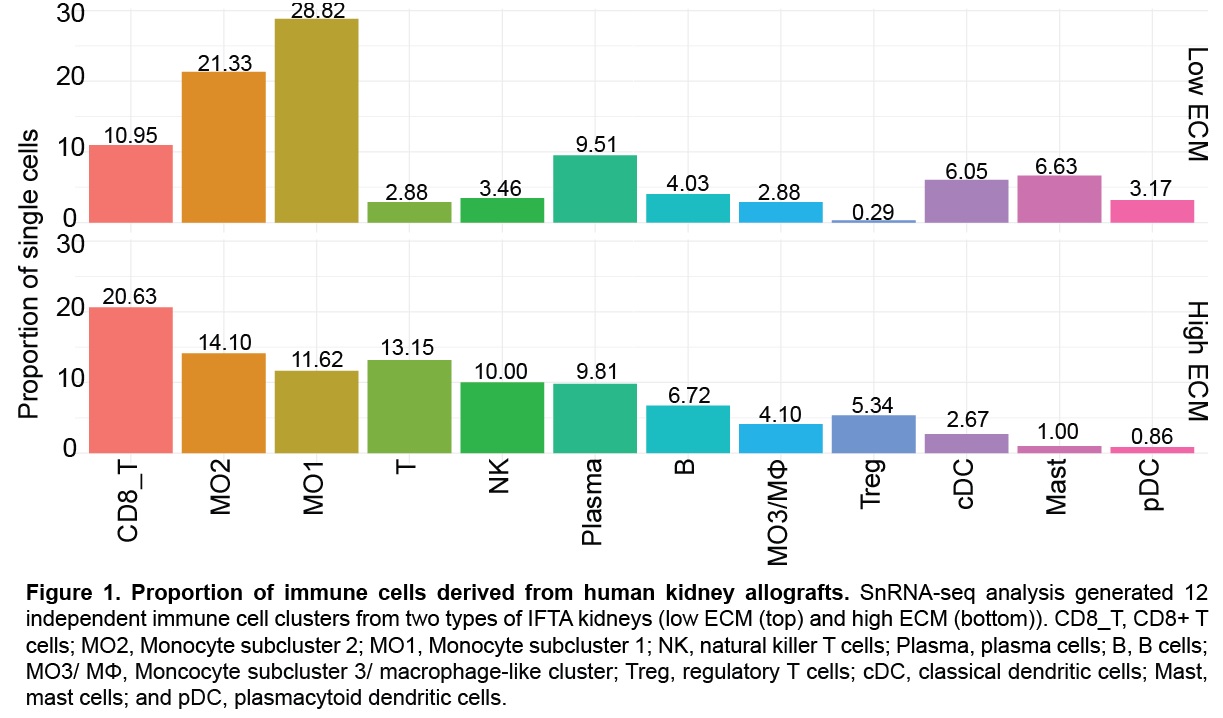
*Results: Differences in extracellular matrix (ECM) production and immune cells led to the identification of two IFTA groups, high and low ECM. A total of 12 immune cell subclusters were generated including B cells, dendritic cells, mast cells, monocytes, plasma cells, and T cells. Low ECM were abundant in monocytes (MO1/MO2), dendritic cells (conventional and plasmacytoid), and mast cells. High ECM accumulated more T cells (CD8+ T cells, NK cells, and Tregs) and B cells. MO1 and MO2 expressed several pro-inflammatory factors including HLA class II antigens (HLA-DRA, HLA-DPA1, HLA-DQA1). Immune cell chimerism was identified, although predominantly of recipient origin. Gene expression analysis also identified a subcluster of resident macrophages in IFTA kidneys, mediating a novel role in fibrosis and tissue repair. Monocyte paracrine signaling highlighted activation of diverse cellular processes and reciprocal anti- and pro-inflammatory signaling.
*Conclusions: To our knowledge, this is the first time the immune cell landscape is described in human kidneys of chronic injury ≥15mos posttranslantation. These findings show that kidney grafts carry immune cells of both donor- and recipient-origin for many years after KT. The differences in immune cell abundance and specific cell types found in fibrotic kidneys may contribute to the activation of the adaptive versus innate response. Assessment of immune responses and signaling is necessary to uncover the dysfunctional pathways related to kidney graft injury and outcomes.
To cite this abstract in AMA style:
McDaniels J, Shetty A, Kuscu C, Kuscu C, Rousselle T, Bardhi E, Drachenberg C, Talwar M, Eason J, Muthukumar T, Maluf D, Mas V. Single Nuclei RNA-Sequencing Identifies Distinct Immune Cell Profile in Kidney Allograft Fibrosis [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/single-nuclei-rna-sequencing-identifies-distinct-immune-cell-profile-in-kidney-allograft-fibrosis/. Accessed March 9, 2026.« Back to 2022 American Transplant Congress