Sequential Analysis of Renal Allograft Rejection at Single Cell Resolution
T. Shi1, C. Castro-Rojas1, A. Burg1, K. Roskin1, J. Rush2, B. Haraldsson2, A. Shields3, R. Alloway3, E. Woodle3, D. Hildeman1
1Immunobiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 2Novartis Pharmaceuticals AG, Basel, Switzerland, 3University of Cincinnati Medical Center, Cincinnati, OH
Meeting: 2021 American Transplant Congress
Abstract number: 257
Keywords: Gene expression, Graft-infiltrating lymphocytes, Kidney transplantation, Rejection
Topic: Basic Science » Lymphocyte Biology: Signaling, Co-Stimulation, Regulation
Session Information
Session Name: Lymphocyte Biology and Tolerance
Session Type: Rapid Fire Oral Abstract
Date: Monday, June 7, 2021
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-6:05pm
Presentation Time: 6:00pm-6:05pm
Location: Virtual
*Purpose: Renal allograft anti-rejection therapies (corticosteroids (CCS) and anti-lymphocyte globulin) have been the primary treatment modalities for over six decades, yet little is known about their effects on graft infiltrating immune cells. Similarly, the understanding of rejection biology occurring under CNIs or new biologics is lacking. Here, we defined the transcriptomic profiles of kidney tissue and infiltrating immune cells and paired urine samples at the time of rejection and after anti-rejection therapy (RejTx).
*Methods: Renal allograft recipients enrolled in a clinical trial of iscalimab, an anti-CD40 mAb, were followed longitudinally if they had rejection. Renal allograft biopsies and paired urine samples were collected at the initial rejection and after RejTx. 5’ single cell RNA sequencing was performed with linked TCR sequencing and clonal analyses.
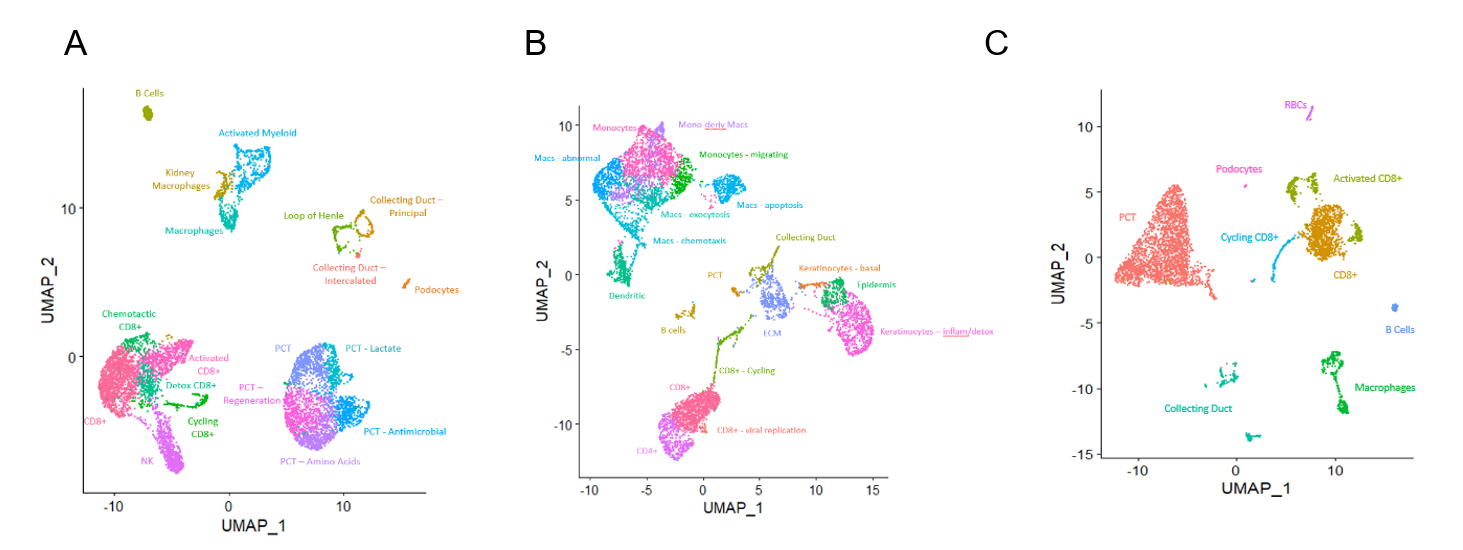
*Results: One patient with a Banff 1B rejection underwent RejTx (CCS, CNI, MMF) and failed to resolve rejection, a process captured across 4 biopsies. Throughout the course of rejection, inflammatory signals in multiple cell types in the graft and urine were observed. The top 7 expanded CD8+ T cell clones persisted for weeks but were eventually displaced by a new set of expanded clones, even in the presence of ongoing rejection. In contrast, another patient followed over 2 biopsies experienced a Banff 1A rejection characterized by a similar inflammatory gene expression across multiple cell types and several expanded CD8+ T cell clones. In this patient, the top 7 expanded clones were all similarly diminished after RejTx. Thus, rejection reversal was associated with a decrease in inflammatory gene expression, but not a rapid change in expanded T cell clones in the graft. Ongoing work will further characterize differential gene expression across multiple cell types in ongoing rejection compared to resolving rejection.
Figure 1: UMAP of patient 1 biopsy (A) and urine (B) and patient 2 biopsy (C), integrated across timepoints.
*Conclusions: 1) Refractory rejection under iscalimab is associated with failure to eliminate dominant TCR clones in the allograft and an emergence of new T cell clones, 2) urine and renal allograft biopsies display similar cell populations, including clonally dominant CD8+ T cells, 3) rejection reversal is associated with decreased inflammatory gene expression in kidney tissue and infiltrating immune cells.
To cite this abstract in AMA style:
Shi T, Castro-Rojas C, Burg A, Roskin K, Rush J, Haraldsson B, Shields A, Alloway R, Woodle E, Hildeman D. Sequential Analysis of Renal Allograft Rejection at Single Cell Resolution [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/sequential-analysis-of-renal-allograft-rejection-at-single-cell-resolution/. Accessed March 2, 2026.« Back to 2021 American Transplant Congress