Multiplexed Droplet Single-Cell Sequencing (mux-seq) Identifies Novel Cell States in Renal Allograft Rejection
UCSF, San Francisco, CA
Meeting: 2022 American Transplant Congress
Abstract number: 922
Keywords: Kidney transplantation, Nephropathy
Topic: Basic Science » Basic Science » 07 - Vascular, Lymphatic, Stromal and Parenchymal Cell Biology
Session Information
Session Name: Vascular, Lymphatic, Stromal and Parenchymal Cell Biology
Session Type: Poster Abstract
Date: Sunday, June 5, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
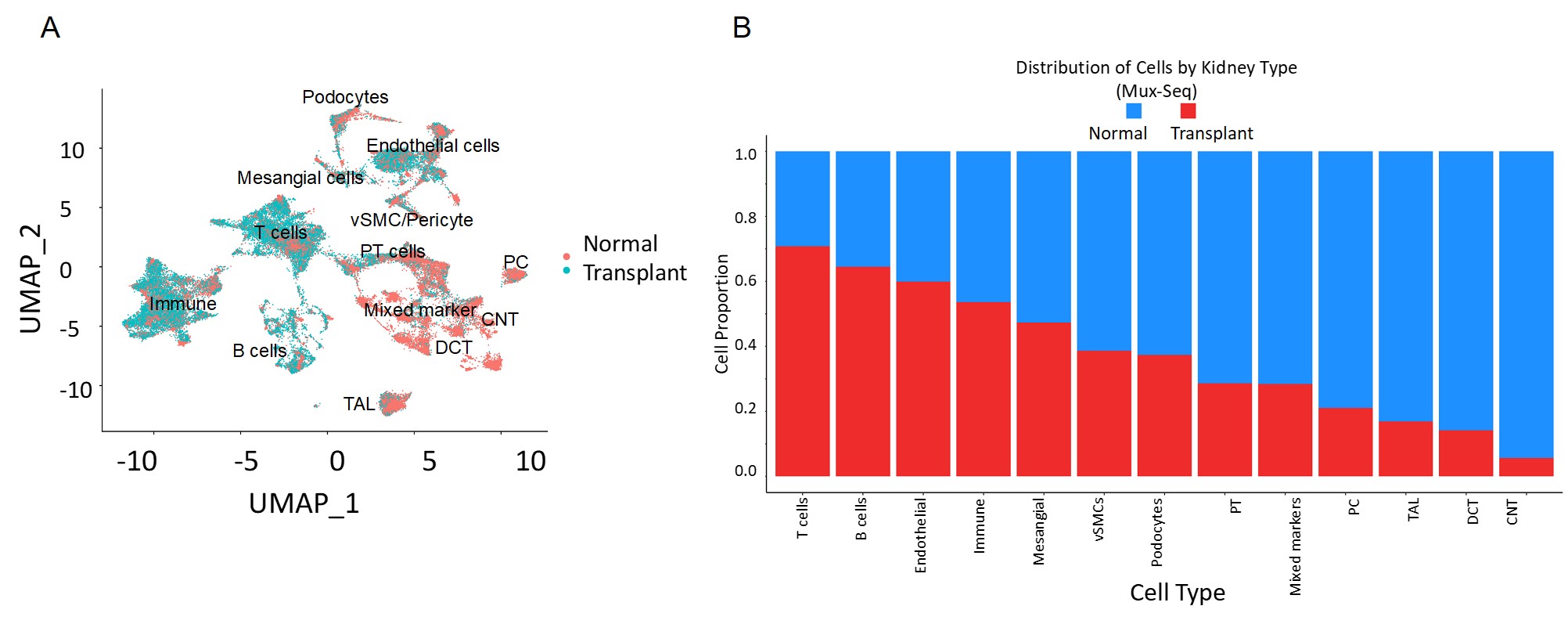
*Purpose: Maintenance of systemic homeostasis by kidney requires the coordinated response of diverse cell types. Use of single-cell RNA sequencing (scRNAseq) for patient samples remains fraught with difficulties. The ability to characterize immune and parenchymal cells during transplant rejection (some of which may be present at very small numbers due to injury) will be invaluable in defining transplant pathologies.
*Methods: Explant tissues from 6 normal and 2 transplant recipients after multiple rejection episodes leading to nephrectomy were pooled for Mux-Seq. Subsequently, a computational tool, Demuxlet was applied for demultiplexing the individual cells. Each sample was also applied individually in single microfluidic run (singleplex). Cell trajectory analysis was conducted to identify evolution of different cell states in rejection.
*Results: We show that data from Mux-Seq correlated highly with singleplex (Pearson coefficient 0.982). Both are able to identify many known kidney cell types including immune cells. Trajectory analysis of proximal tubule and endothelial cells demonstrates separation between healthy and injured kidney from transplant explant suggesting various stages of differentiation.
*Conclusions: Mux-Seq has the potential to minimize experimental batch biases and variations and reduce assay costs. This study provides the technical groundwork for understanding the pathogenesis of alloimmune injury and the evolution of renal proximal tubule and endothelial cell states in acute rejection, uncovering possible new targets for drug design
To cite this abstract in AMA style:
Rashmi P, Sur S, Sigdel T, Boada P, Sarwal M. Multiplexed Droplet Single-Cell Sequencing (mux-seq) Identifies Novel Cell States in Renal Allograft Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/multiplexed-droplet-single-cell-sequencing-mux-seq-identifies-novel-cell-states-in-renal-allograft-rejection/. Accessed March 10, 2026.« Back to 2022 American Transplant Congress