Multiplexed Droplet Single-Cell RNA-Sequencing for Cellular Profiling of Transplant Rejection
A. Schroeder, T. Sigdel, I. Damm, S. Sur, G. Hartoularos, J. Ye, M. Sarwal
Department of Surgery, UCSF, San Francisco, CA
Meeting: 2019 American Transplant Congress
Abstract number: A10
Keywords: Endothelial cells, Graft-infiltrating lymphocytes, Kidney, Nephrectomy
Session Information
Session Name: Poster Session A: Acute Rejection
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: Multiplexed droplet single-cell RNA-sequencing (mdscRNA-seq) has been utilized across many tissue types for fast and paralleled transcriptomic profiling. However, maintaining sample identity for differential expression analysis has been an issue. A novel computational tool, demuxlet, has been developed that harnesses natural genetic variation to preserve sample identity. Here we apply this tool for kidney cell type identification to better understand the pathophysiology of the rejecting kidney and intragraft cellular infiltrates.
*Methods: Cells isolated from 4 Cryostor preserved kidney biopsy samples (2 non-rejection, 2 transplant rejection) were isolated and pooled (multiplexed) for mdscRNA-sequencing using the 10X Genomics platform. The Demuxlet tool was applied to demultiplex the samples via SNP array. Kidney cell type profiling and quality control analysis were performed using the R package Seurat.
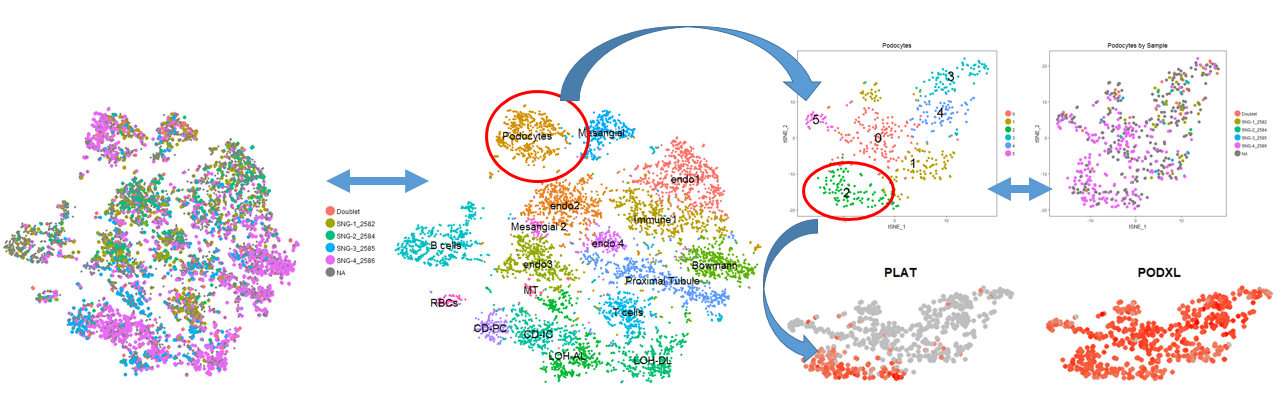
*Results: Using 25-50 SNPs per kidney sample was sufficient for Demuxlet to precisely identify the sample origin of >90% of droplet singlets as well as doublets (Figure). 17-20 cell types were identified, including glomerular, tubular, and immune cells. Unsupervised clustering of individual cell types was done to explore potentially novel cell subtype marker genes. The figure displays a workflow for the investigation of novel cellular subclusters.
*Conclusions: Using Demuxlet, dscRNA-seq is made more efficient by removing intersample batch effects, decreasing tissue and cell quantity needed, and lowering assay cost and time many fold. We have shown that this is an effective tool for preserving sample identity for downstream analysis for differential gene and cell-types across samples as well as discovery of novel kidney cell markers and cell subtypes in transplant rejection.
To cite this abstract in AMA style:
Schroeder A, Sigdel T, Damm I, Sur S, Hartoularos G, Ye J, Sarwal M. Multiplexed Droplet Single-Cell RNA-Sequencing for Cellular Profiling of Transplant Rejection [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/multiplexed-droplet-single-cell-rna-sequencing-for-cellular-profiling-of-transplant-rejection/. Accessed March 5, 2026.« Back to 2019 American Transplant Congress