Molecular Profiling of the Injury-Repair Response in Early Subclinical Inflammation
UAB School of Medicine, Birmingham.
Meeting: 2018 American Transplant Congress
Abstract number: 60
Keywords: Gene expression, Kidney transplantation, Pediatric
Session Information
Session Name: Concurrent Session: Kidney: Pediatrics - 1
Session Type: Concurrent Session
Date: Sunday, June 3, 2018
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:42pm-3:54pm
Presentation Time: 3:42pm-3:54pm
Location: Room 3AB
Purpose: Surveillance biopsies can detect subclinical T-cell mediated rejection (SC-TCMR) or subclinical borderline rejection (SC-B-TCMR) in kidney transplants with normal/stable creatinine. Some consider SC-B-TCMR to be mild allograft injury that does not require treatment. However, we recently showed that pediatric kidney recipients with SC-B-TCMR and SC-TCMR have equally poor outcomes compared to those with no major surveillance abnormalities (NOMOA). We hypothesized that molecular profiling could identify shared injury-repair responses that distinguish SC-B-TCMR and SC-TCMR from NOMOA.
Methods: Nested case-control study (n=35) within a cohort of 103 consecutive pediatric kidney transplant recipients with surveillance biopsies at < 6 months post-transplant. Cases (n=18) had subclinical inflammation defined as SC-B-TCMR (n=11) or SC-TCMR (n=7) by Banff 2013 criteria. Controls (n=17) had NOMOA plus 100% rejection-free graft survival at 5 years. We isolated RNA from formalin-fixed, paraffin-embedded surveillance biopsies residual to clinical use, and directly counted 770 genes in 13 injury-repair response pathways using the Nanostring platform. We used linear regression and unsupervised hierarchical clustering to compare gene counts by histology, and to group biopsies with similar molecular profiles.
Results: SC-B-TCMR and SC-TCMR were molecularly similar with no differentially expressed genes between groups. Comparisons of pathway scores showed that 8/13 injury-repair pathways were similarly expressed between SC-B-TCMR and SC-TCMR yet differentially expressed from NOMOA (Figure). 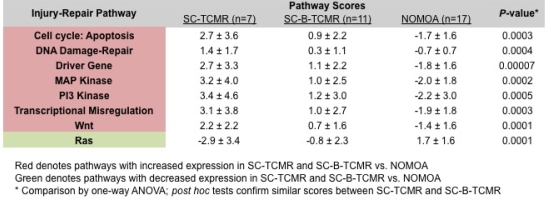
This was confirmed by unsupervised clustering that divided all 35 biopsies into 2 distinct molecular groups: Group 1 (n=15) included 87% with SC-B-TCMR or SC-TCMR, whereas Group 2 (n=20) included 75% with NOMOA (P=0.001).
Conclusions: SC-B-TCMR and SC-TCMR had similar molecular profiles of allograft injury that were significantly distinct from NOMOA. These shared molecular profiles may partially explain why SC-B-TCMR and SC-TCMR have poor outcomes in children despite histological differences in allograft injury.
CITATION INFORMATION: Seifert M., Della Manna D., Liu X., Rosenblum F., Yanik M., Yang E., Pollock J., Mannon R. Molecular Profiling of the Injury-Repair Response in Early Subclinical Inflammation Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Seifert M, Manna DDella, Liu X, Rosenblum F, Yanik M, Yang E, Pollock J, Mannon R. Molecular Profiling of the Injury-Repair Response in Early Subclinical Inflammation [abstract]. https://atcmeetingabstracts.com/abstract/molecular-profiling-of-the-injury-repair-response-in-early-subclinical-inflammation/. Accessed February 20, 2026.« Back to 2018 American Transplant Congress
