Molecular Profiles of Renal Allograft Rejection.
Internal Medicine/Renal Medicine, University of Illinois at Chicago, Chicago, Il.
Meeting: 2016 American Transplant Congress
Abstract number: 33
Keywords: Gene expression, Kidney transplantation, Rejection
Session Information
Session Name: Concurrent Session: Kidney Acute Cellular Rejection: Clinical Outcomes and Pathological Characteristics
Session Type: Concurrent Session
Date: Sunday, June 12, 2016
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:42pm-3:54pm
Presentation Time: 3:42pm-3:54pm
Location: Ballroom C
Introduction: The gold standard for the diagnosis of allograft rejection is the Banff Classification, yet it shows substantial variability among pathologists. Using molecular signatures of differentially expressed genes, we identified different subtypes of rejection. The development of molecular signatures of subsets of Banff classifications of allograft rejection could increase diagnostic precision. Methods: Microarray expression data of kidney transplant biopsies ( NCBI GEO) was analyzed using R language. Differential gene expression of log2 fold change was determined by student's TTEST (p < 0.05) with correction for multiple testing by false discovery rate (fdr <0.05). Different subtypes of antibody-mediated (ABMR), T-cell-mediated (TCMR) and borderline (Bord) rejection were identified by hierarchical clustering analysis and evaluated by connectivity, Dunn and Silhouette index. Differentially expressed KEGG pathways were identified with CPDB resources to analyze biological functions. Results: We identified multiple subtypes of TCMR, ABMR and Bord rejection based on molecular signatures. We previously reported differential expression and functions in 3 subtypes of TCMR. In this analysis, we detected a core group of genes plus subtype specific unique genes in the 3 Banff classifications. The core and the ABMR02 groups expressed immune functions .Interestingly, the TCMR01 and ABMR01 unique gene subtypes predominantly involved metabolic functions .Tryptophan metabolism was noted to be uniquely expressed in the TCMR01 and Borderline01 group. The metabolism of this A.A. has been highly implicated with IFN-gamma secretion. 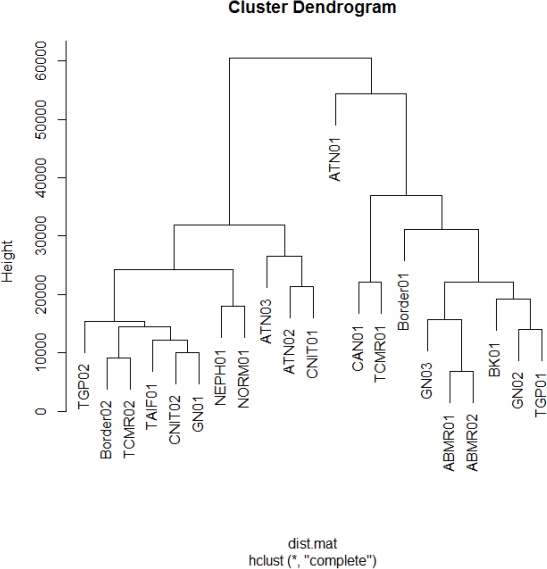
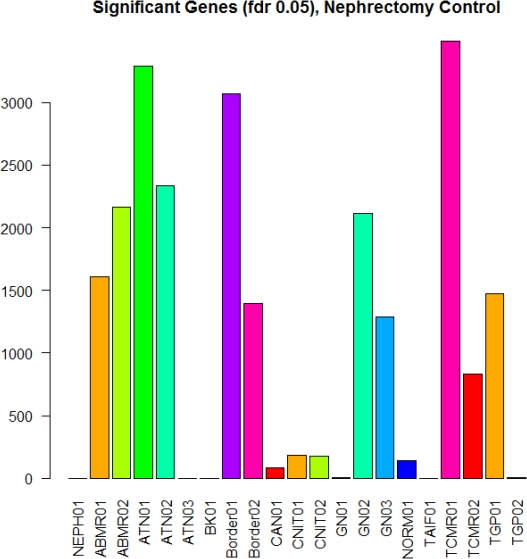 Conclusion: A distinct metabolic profile was noted in the ABMR01, TCMR01 and Bord01 subtypes. This "metabolic signature" may be considered a valuable tool in establishing the diagnosis of rejection and developing novel therapeutic strategies.
Conclusion: A distinct metabolic profile was noted in the ABMR01, TCMR01 and Bord01 subtypes. This "metabolic signature" may be considered a valuable tool in establishing the diagnosis of rejection and developing novel therapeutic strategies.
CITATION INFORMATION: Hajjiri Z, Finn P, Perkins D. Molecular Profiles of Renal Allograft Rejection. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Hajjiri Z, Finn P, Perkins D. Molecular Profiles of Renal Allograft Rejection. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/molecular-profiles-of-renal-allograft-rejection/. Accessed February 22, 2026.« Back to 2016 American Transplant Congress
