Molecular Associations in Kidney Transplant Biopsies in the Trifecta Study Indicate That at Least Three Distinct Processes Release Dd-cfDNA: ABMR, TCMR, and Injury
1Alberta Transplant Applied Genomics Centre, Edmonton, AB, Canada, 2Natera Inc., San Carlos, CA, 3., ., AB, Canada
Meeting: 2022 American Transplant Congress
Abstract number: 214
Keywords: Biopsy, Kidney transplantation, Rejection
Topic: Clinical Science » Kidney » 34 - Kidney: Acute Cellular Rejection
Session Information
Session Name: Kidney: Acute Cellular Rejection
Session Type: Rapid Fire Oral Abstract
Date: Monday, June 6, 2022
Session Time: 3:30pm-5:00pm
 Presentation Time: 4:40pm-4:50pm
Presentation Time: 4:40pm-4:50pm
Location: Hynes Ballroom A
*Purpose: In the Trifecta study (ClinicalTrials.gov # NCT04239703 53), elevated plasma dd-cfDNA is associated with molecular rejection in kidney transplant indication biopsies, particularly active ABMR (JASN, in press 2021). However, many biopsies with active TCMR and injury also had elevated dd-cfDNA. This could reflect undetected ABMR, or direct effects of TCMR and injury on dd-cfDNA. The present analysis of 426 Trifecta biopsies sought to determine whether dd-cfDNA elevation in TCMR and no rejection represents undetected ABMR or release of dd-cfDNA by other disease states.
*Methods: %dd-cfDNA was measured by Natera (Prospera) and MMDx (genome-wide mRNA expression) at ATAGC. We examined %dd-cfDNA correlations with molecular findings within biopsy MMDx groups: ABMR (+ possible ABMR) 121, TCMR (+ possible TCMR) 33, No rejection 246, and 26 mixed rejection.
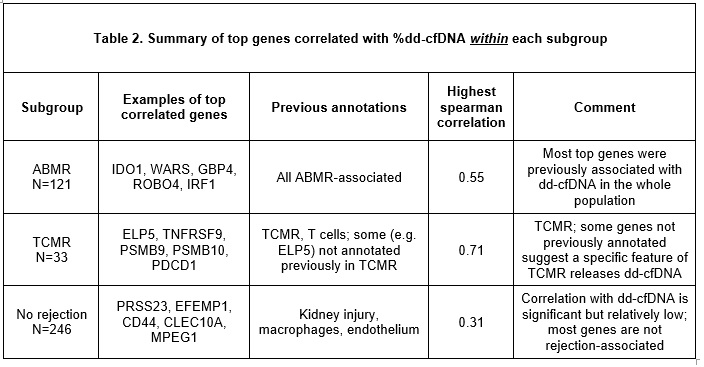
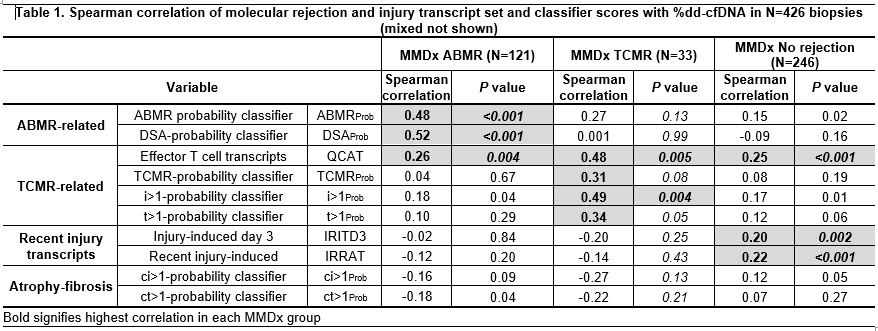
*Results: As previously reported, %dd-cfDNA was elevated in rejection: TCMR 2.36; ABMR 2.70; no rejection 0.64. We studied correlations of gene sets, classifiers, and single genes with elevations of %dd-cfDNA within subgroups. In Table 1, the highest %dd-cfDNA correlations with classifier and transcript set scores differed among subgroups: ABMR-related scores in ABMR, TCMR-related scores in TCMR, and recent injury scores in No rejection. Atrophy-fibrosis had little association. Table 2 summarizes the top genes by Spearman correlation (SCC) within each subgroups. In ABMR, the correlations were ABMR transcripts, with top SCC=0.55. In TCMR, SCCs were highest (0.71) and the top genes included typical TCMR transcripts (e.g. TNFSF9, PDCD1), but some not previously annotated (e.g. ELP5). In No rejection, the top genes included kidney injury and macrophage genes, but with a relatively low SCC=0.31.
*Conclusions: Associations with gene set and classifier scores and expression of single genes reveals that at least three different conditions release dd-cfDNA: ABMR, TCMR, and recent parenchymal injury. Thus, dd-cfDNA release in TCMR and injury is not due to undetected ABMR.
To cite this abstract in AMA style:
Halloran PF, Reeve J, Madill-Thomsen KS, Demko Z, Prewett A, Billings P. Molecular Associations in Kidney Transplant Biopsies in the Trifecta Study Indicate That at Least Three Distinct Processes Release Dd-cfDNA: ABMR, TCMR, and Injury [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/molecular-associations-in-kidney-transplant-biopsies-in-the-trifecta-study-indicate-that-at-least-three-distinct-processes-release-dd-cfdna-abmr-tcmr-and-injury/. Accessed February 28, 2026.« Back to 2022 American Transplant Congress