Mapping Heterogeneity in Antibody-Mediated Rejection Using Molecular Phenotypes and Archetype Analysis.
1Alberta Transplant Applied Genomics Centre, Edmonton, AB, Canada
2Medicine, University of Alberta, Edmonton, AB, Canada.
Meeting: 2016 American Transplant Congress
Abstract number: 119
Keywords: Biopsy, Kidney transplantation, Rejection
Session Information
Session Name: Concurrent Session: Kidney AMR: Making the Diagnosis
Session Type: Concurrent Session
Date: Sunday, June 12, 2016
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:42pm-4:54pm
Presentation Time: 4:42pm-4:54pm
Location: Veterans Auditorium
Archetype analysis is a method for identifying new relationships among samples without subdividing them into classes, finding a limited number of archetypical cases and calculating the relationship of every sample to each archetype. We applied this method to molecular ABMR, using microarray and conventional data from 1208 indication biopsies from prospective multicenter studies in Europe and North America (Clinical trials.govNTC1299168). Each sample had scores assigned by molecular classifiers trained on its histologic ABMR diagnosis (ABMR score) or its histologic features (ptc-, g-, or cg-lesions). The analysis indicated that 5 archetypes explained the variation in the ABMR features, and that all biopsies had relationships to all five archetypes: 1. late cg-rich with less ptc/g than full house; 2. “full house” highly active ABMR; 3. early ABMR without cg features; 4. A form of TCMR that shared some ABMR features, but had unexpectedly low hyalinosis for time and probably reflected non-adherence; 5. Biopsies with no ABMR-related molecular features. Biopsies were divided into clusters based on their strongest archetype relationships 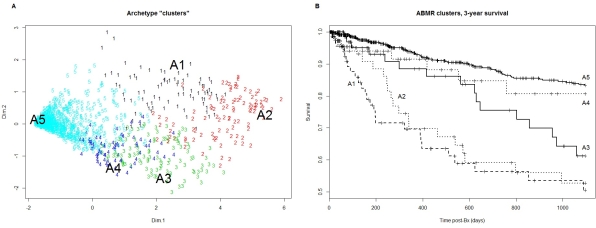 . Risk of subsequent failure was highest in the late cg-rich (#1) and full house (#2) clusters, but after a delay was also increased in the early ABMR biopsies without cg
. Risk of subsequent failure was highest in the late cg-rich (#1) and full house (#2) clusters, but after a delay was also increased in the early ABMR biopsies without cg  . This method indicates that many biopsies that may be misclassified by histology
. This method indicates that many biopsies that may be misclassified by histology 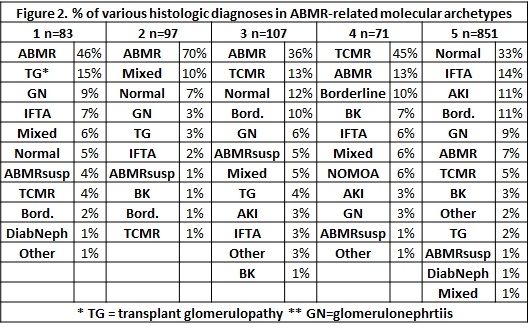 , including histologic ABMR lacking molecular features, histologic TCMR misclassified as ABMR and vice versa, etc. The results raise the possibility of better disease classification and risk prediction when new molecular methods of assessment are combined with novel methods for discovering relationships among biopsies while retaining the uniqueness of each biopsy.
, including histologic ABMR lacking molecular features, histologic TCMR misclassified as ABMR and vice versa, etc. The results raise the possibility of better disease classification and risk prediction when new molecular methods of assessment are combined with novel methods for discovering relationships among biopsies while retaining the uniqueness of each biopsy.
CITATION INFORMATION: Reeve J, Halloran P. Mapping Heterogeneity in Antibody-Mediated Rejection Using Molecular Phenotypes and Archetype Analysis. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Reeve J, Halloran P. Mapping Heterogeneity in Antibody-Mediated Rejection Using Molecular Phenotypes and Archetype Analysis. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/mapping-heterogeneity-in-antibody-mediated-rejection-using-molecular-phenotypes-and-archetype-analysis/. Accessed March 5, 2026.« Back to 2016 American Transplant Congress
