Is the Epitope Mismatch Load Enough to Identify HLA-DQ Mismatches with a Low Risk for De Novo DSA Development?
1Terasaki Research Institute, Los Angeles, CA, 2Mayo Clinic, Rochester, MN, 3Mayo Clinic, Phoenix, AZ, 4East Carolina University, Greenville, NC, 5Henry Ford Transplant Institute, Detroit, MI
Meeting: 2020 American Transplant Congress
Abstract number: A-282
Keywords: Epitopes, HLA antibodies, HLA matching, Kidney transplantation
Session Information
Session Name: Poster Session A: Histocompatibility and Immunogenetics
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: The development of de novo donor-specific anti-HLA antibodies (dnDSA) remains a major risk factor for poor long-term kidney allograft outcomes. In particular, anti-HLA-DQ dnDSAs, which are resistant to treatment, have the highest association with poor allograft outcomes. The purpose of this study is to evaluate whether HLA-DQ epitope matching from serological HLA-DQ typing may identify HLA-DQ mismatches (MMs) with a lower risk for anti-HLA-DQ dnDSA development in a large multi-center cohort.
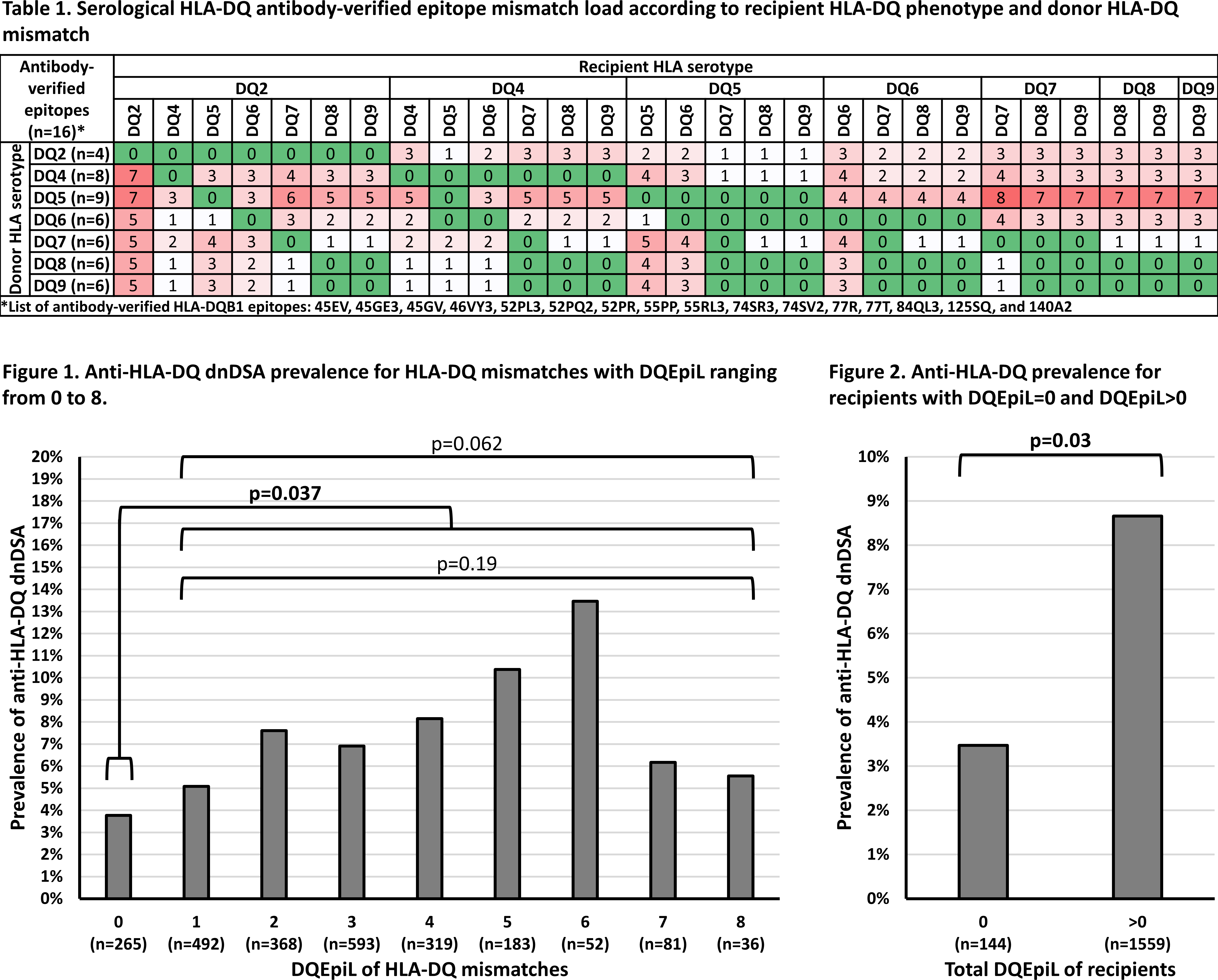
*Methods: We retrospectively studied 1703 HLA-DQ mismatched kidney recipients, without preformed DSA, that were transplanted between 2007 and 2016 at four U.S. centers without preformed DSA. HLA-DQ serological epitope mismatch load (DQEpiL) was assessed using Table 1, where only antibody-verified HLA-DQB1 epitopes present on any HLA-DQB1 molecule of a same HLA-DQ serology are considered.
*Results: Among the 1703 recipients, we observed a total of 2389 HLA-DQ MMs, of which 163 induced a dnDSA. There was no significant difference in the prevalence of dnDSA between DQ MMs with DQEpiL from 0 to 8 (p=0.065). However, the prevalence of dnDSA was significantly higher for HLA-DQ MMs with DQEpiL>0 compared to HLA-DQ MMs with DQEpiL=0 (7.2% vs. 3.8%, respectively) (p=0.037), but there was no significant difference in the prevalence of dnDSA among DQ MMs with DQEpiL>0 (p=0.19) (Figure 1). Last, stratifying recipients based on total DQEpiL=0 identified 144 (8.5%) recipients, which had a significantly lower prevalence of anti-HLA-DQ dnDSA compared to recipients with a total DQEpiL>0 (3.5% vs. 8.7%, respectively) (p=0.030) (Figure 2).
*Conclusions: This study shows that, in contrast to the level of DQEpiL, HLA-DQ MMs with DQEpiL=0 induce dnDSA significantly less often than HLA-DQ MMs with DQEpiL>0. Moreover, the stratification of recipients based on total DQEpiL=0 significantly lowers the prevalence of anti-HLA-DQ dnDSA. Serological HLA-DQ epitope matching may be a strategy to reduce the development of dnDSA, although further studies are warranted.
To cite this abstract in AMA style:
Jucaud V, Stegall M, Schinstock C, Heilman R, Rebellato L, Samaniego-Picota M, Leeser D, Gandhi M, Beaumont J, Everly M. Is the Epitope Mismatch Load Enough to Identify HLA-DQ Mismatches with a Low Risk for De Novo DSA Development? [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/is-the-epitope-mismatch-load-enough-to-identify-hla-dq-mismatches-with-a-low-risk-for-de-novo-dsa-development/. Accessed February 19, 2026.« Back to 2020 American Transplant Congress