Informatics Tools for Mapping Molecular HLA Typing Data to UNOS Antigen Equivalencies
1Tulane University School of Medicine, New Orleans
2Cedars-Sinai Heart Institute, Los Angeles
3Baylor Scott and White, Temple
4National Marrow Donor Program / Be The Match, Minneapolis.
Meeting: 2018 American Transplant Congress
Abstract number: B105
Keywords: Allocation, HLA antigens, Kidney transplantation, Major histocompatibility complex (MHC)
Session Information
Session Name: Poster Session B: Kidney Deceased Donor Allocation
Session Type: Poster Session
Date: Sunday, June 3, 2018
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall 4EF
Introduction:
Molecular HLA typing must be converted to antigen equivalencies for entry into the UNOS database, UNet. Maintaining an up-to-date mapping table for this purpose has so far been deemed infeasible by UNOS. To automate the conversion process for histocompatibility laboratories and researchers, we aimed to develop a mapping table and supporting informatics tools that would handle both unambiguous and ambiguous molecular HLA typings.
Methods:
Data sources included a OPTN policy document for UNOS antigen mapping guidelines and equivalency tables, an IMGT/HLA antigen mapping table maintained by WHO and the World Marrow Donor Association (WMDA) and the 2008 HLA Dictionary. Mapping of ambiguous HLA typings utilizes published National Marrow Donor Program (NMDP) allele frequencies from 26 US populations. Validation of antigen mapping was done by mapping NMDP high resolution frequencies to antigens then comparing with UNOS calculated panel reactive antibody (CPRA) antigen frequencies using a normalized similarity index.
Results:
We present a UNOS antigen mapping table for 16093 HLA alleles of the A, B, C, DRB1, DRB3/4/5, DQA1, and DQB1 loci from the v.3.30.0 release of the IMGT/HLA database. A web application and microservices to map HLA typing results were also developed for use by histocompatibility labs (Available at http://www.transplanttoolbox.org). Similarity scores between the NMDP reference panel and the UNOS CPRA reference panel for A, C, B, DR and DQ antigen frequencies for 4 US broad population categories (Caucasians, Hispanics, African Americans and Asian/Pacific Islanders) ranged from 0.85 to 0.97, indicating that the alleles were correctly mapped to antigens.
Conclusion:
The table provides a standardized reference for antigen mapping that can be automatically updated to incorporate new alleles added to IMGT/HLA. Informatics tools for managing molecular HLA typing data may improve accuracy and reduce the manual effort needed to enter data into UNet. 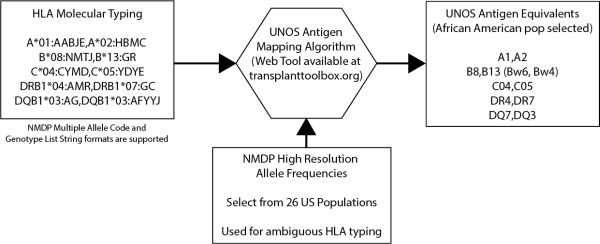
CITATION INFORMATION: Kaur N., Kransdorf E., Pando M., Maiers M., Gragert L. Informatics Tools for Mapping Molecular HLA Typing Data to UNOS Antigen Equivalencies Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Kaur N, Kransdorf E, Pando M, Maiers M, Gragert L. Informatics Tools for Mapping Molecular HLA Typing Data to UNOS Antigen Equivalencies [abstract]. https://atcmeetingabstracts.com/abstract/informatics-tools-for-mapping-molecular-hla-typing-data-to-unos-antigen-equivalencies/. Accessed February 19, 2026.« Back to 2018 American Transplant Congress
