Development and Validation of an Integrative DD-CFDNA System to Predict Allograft Rejection: A Population Based Study
Paris Translational Center for Organ Transplantation, Paris, France
Meeting: 2022 American Transplant Congress
Abstract number: 563
Keywords: Biopsy, Kidney, Rejection
Topic: Basic Science » Basic Clinical Science » 17 - Biomarkers: Clinical Outcomes
Session Information
Session Name: Biomarkers: Clinical Outcomes II
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 7, 2022
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:40pm-5:50pm
Presentation Time: 5:40pm-5:50pm
Location: Hynes Ballroom A
*Purpose: Post-transplantation patient care requires development and validation of non-invasive biomarkers to improve allograft monitoring and prevention from unnecessary and costly biopsies. Reports have suggested the association of donor derived cell-free DNA (dd-cfDNA) with allograft rejection. However, there is no proof of its added value on standard of care in large, unselected and deep phenotyped cohort.
*Methods: We enrolled 992 kidney transplant recipients having concomitant evaluation of allograft histology, anti-HLA DSA and functional parameters between April 2013 and December 2020, representing 1,210 biopsies. dd-cfDNA was measured in plasma at the time of the biopsy. The cohort was divided into a derivation cohort (N=637 evaluations) and a validation cohort (N=573 evaluations). Diagnoses were performed with strict application of Banff 2019 criteria. In the derivation cohort, 100 AMR, 14 TCMR and 7 mixed rejections occurred. Parameters associated with rejection were assessed using uni- and multivariable logistic regression. We then developed a risk model using independently associated variables. This model was validated in the validation cohort.
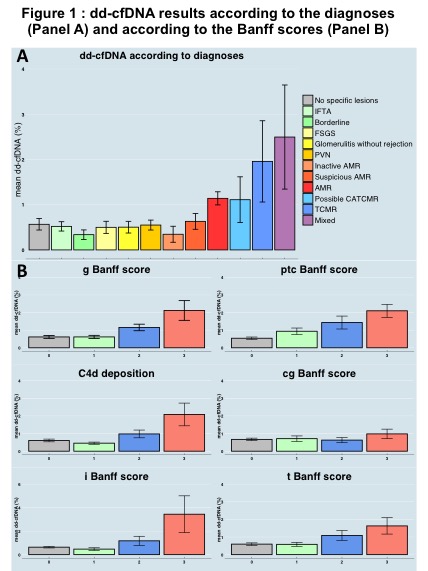
*Results: Higher levels of dd-cfDNA were observed for AMR and TCMR or both compared to other diagnoses (fig.1A). dd-cfDNA incrementally increased with Banff acute lesions without significant increase for chronic lesions (fig.1B). In multivariable analysis, the variables independently associated with rejection were anti-HLA DSA (p<0.0001), dd-cfDNA (p<0.0001), eGFR (p=0.018), recent context of rejection (p<0.0001), and kidney allograft instability (p=0.013). Discrimination of the model without the dd-cfDNA was 0.769 and 0.827 with its inclusion, showing its added value. In the validation cohort, the dicrimination of the model without the dd-cfDNA was 0.786 and 0.824 with its inclusion, showing its added value in the validation cohort.
*Conclusions: We here demonstrate the independent and added value of dd-cfDNA in addition to conventional features to predict rejection. This first integrative system shows improved performance for patient monitoring and could help physicians in decision-making process.
To cite this abstract in AMA style:
Aubert O, Brousse R, Gueguen J, Racape M, Lefaucheur C, Loupy A. Development and Validation of an Integrative DD-CFDNA System to Predict Allograft Rejection: A Population Based Study [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/development-and-validation-of-an-integrative-dd-cfdna-system-to-predict-allograft-rejection-a-population-based-study/. Accessed March 9, 2026.« Back to 2022 American Transplant Congress