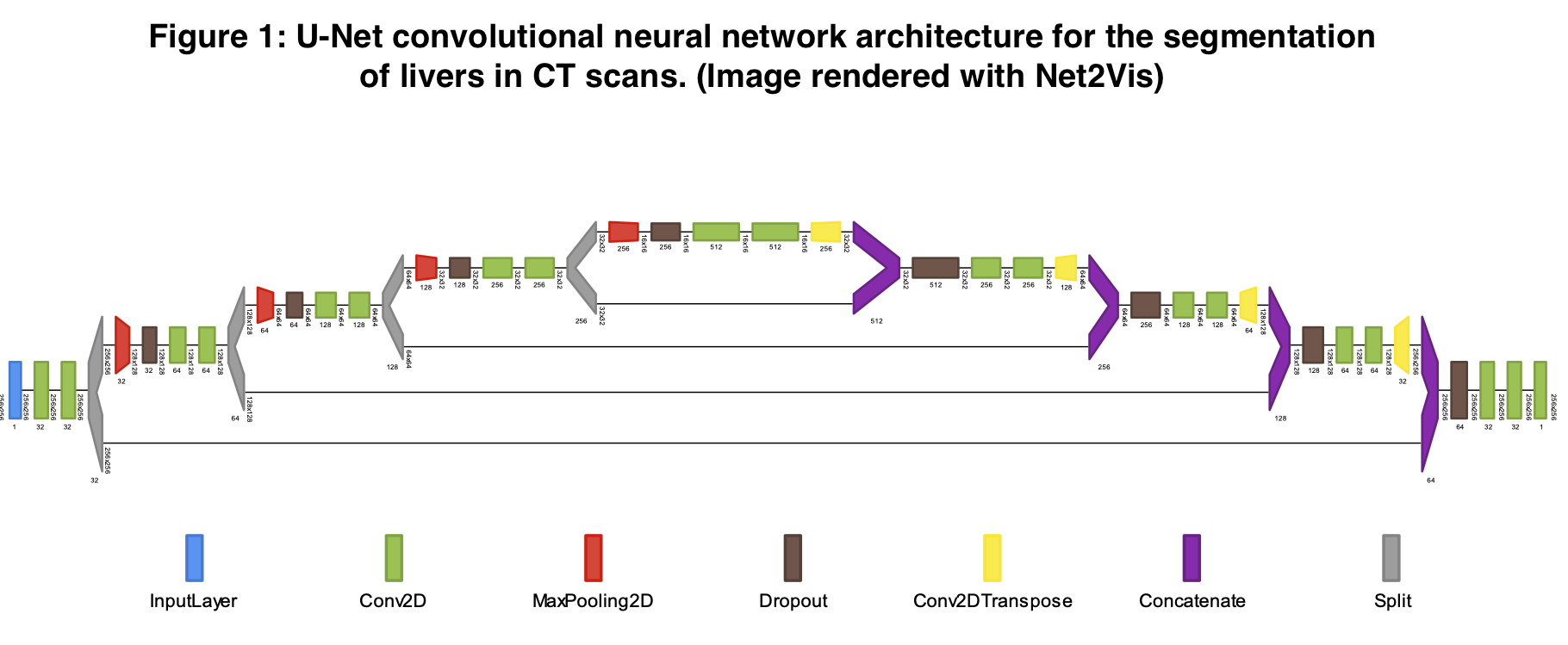
Deep Learning Based Automatic Liver Volume Estimation and Segmentation via U-net Convolutional Neural Network
1InformAI, Houston, TX, 2Baylor College of Medicine, Houston, TX, 3Houston Methodist, Houston, TX
Meeting: 2021 American Transplant Congress
Abstract number: LB 84
Keywords: Liver
Topic: Clinical Science » Liver » Liver: Large Data and Artificial Intelligence
Session Information
Session Name: Liver: Large Data and Artificial Intelligence
Session Type: Poster Abstract
Session Date & Time: None. Available on demand.
Location: Virtual
*Purpose: To create a deep learning model to accurately perform abdominal organ segmentation and volumetric measurements on existing donor and recipient imaging for size matching.
*Methods: The U-Net deep learning image processing algorithm has previously demonstrated success at accurately sectioning out relevant features from standardized medical images. We implemented this method to identify the borders of livers used in solid organ transplantation. Using publicly available CT DICOM scan images from the Liver Tumor Segmentation Challenge (2017) and the 3D Image Reconstruction for Comparison of Algorithm Database (2015) consortiums, we accessed and harmonized 151 abdominal computed tomography scans. For each 3D CT scan, a board-certified radiologist had previously annotated the margins of the liver within the abdominal cavity. We split these data into 64% training, 16% validation, and 20% test sets. After training our U-Net system on these data, we measured the accuracy with which the model identified livers using a Sørenson-Dice coefficient, which measures degree of overlap between predictions and the ground truth. After identifying the borders of the liver with U-Net in 3-dimensional voxels, we multiplied the summed voxels contained in the liver by the X and Y dimensions as well as the CT slice thickness to reconstruct a full organ volume estimate.
*Results: The U-Net model was trained against the Radiologist sectioned CT liver images contained in the training and validation datasets and performance of the model was evaluated against the test dataset. The U-Net model was able to accurately identify the border of the liver with a Dice coefficient of 0.968 in the validation set. The Dice coefficient represents the degree of predicted versus observed overlap and can be interpreted as a 96.8% overlap accuracy. Our calculated volume estimates differed from the Radiologist identified standard liver tracings with a percent error of 4.54% + a standard deviation of 4.49% when testing on the test set.
*Conclusions: Precise volume estimation of livers from CT scans is accurate using a U-Net deep learning architecture. Appropriately deployed, a U-Net algorithm is precise and quick, making it suitable for incorporation into the pre-transplant clinician decision making workflow. Side by side recipient and donor volumetric measurements can greatly facilitate size matching and prevent inefficiencies in organ allocation and reduce discard. This is especially important in pediatric populations where size matching can be difficult to estimate.
To cite this abstract in AMA style:
Marlatt B, Pettit R, Havelka J, Corr SJ, Rana A. Deep Learning Based Automatic Liver Volume Estimation and Segmentation via U-net Convolutional Neural Network [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/deep-learning-based-automatic-liver-volume-estimation-and-segmentation-via-u-net-convolutional-neural-network/. Accessed March 2, 2026.« Back to 2021 American Transplant Congress