Characterization of Hepatocytes During Cold Static Preservation at Single Cell Resolution
1Transplant Research Institute, James D. Eason Transplant Institute, Department of Surgery, The University of Tennessee Health Science Center, Memphis, TN, 2James D. Eason Transplant Institute, Department of Surgery, The University of Tennessee Health Science Center, Memphis, TN, 3University of Tennessee/Methodist Transplant Institute, Memphis, TN
Meeting: 2022 American Transplant Congress
Abstract number: 1306
Keywords: Gene expression
Topic: Basic Science » Basic Science » 16 - Biomarkers: -omics and Systems Biology
Session Information
Session Name: Biomarkers: -omics and Systems Biology
Session Type: Poster Abstract
Date: Monday, June 6, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
*Purpose: Hepatocytes together with liver sinusoidal endothelial cells (LSEC) are the most affected cells during cold preservation of donor liver. Necrotic and apoptotic hepatocytes release HMGB1 which can initiate sterile inflammation and further result in delayed organ function or rejection. Here, we aim to understand the molecular and cellular mechanism of hepatocyte injury in liver allografts at single cell resolution to identify potential intervention mechanisms.
*Methods: We obtained needle biopsy samples from three patients at three different time points; procurement (before cross-clamping and flushing) (LO), pre-implantation (L1) and post-transplantation (L2). Due to technical limitations of proper tissue dissociation, we isolated nuclei from our samples. We used droplet-based 10X Genomics Chromium Platform and run the 3’ kit. Feature-barcode matrix data was generated by CellRanger5.0 and clusters analyses were performed by the combination of high-throughput and hyperparameter approaches along with unsupervised “Seurat” package.
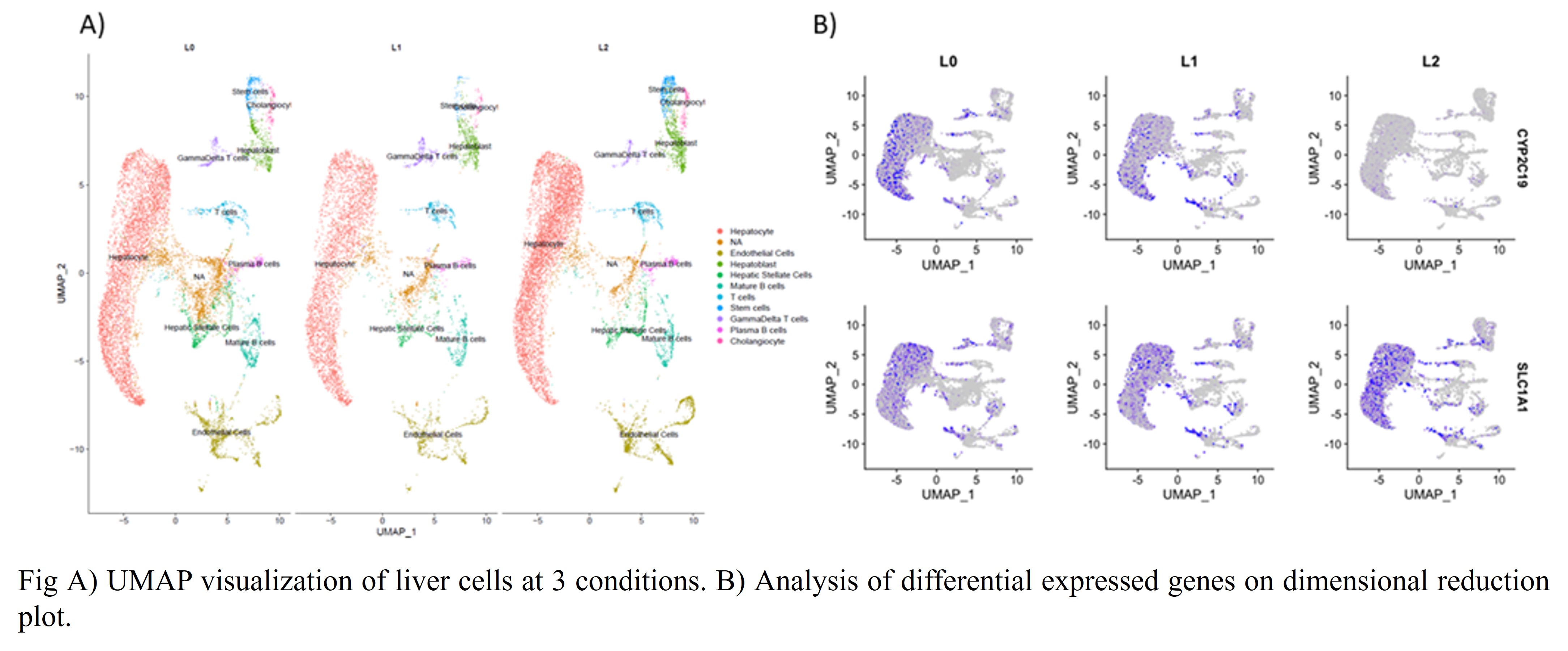
*Results: We obtained ~30,000 cells in total with an average >2,000 genes per cell. We integrated 9 different datasets by Seurat pipeline and identified 19 different clusters. We identified same types of liver cells that have been identified before via single-cell approach. There is no significant change in the total number of hepatocytes and LSECs based on the conditions (L0 vs L1 vs L2) (Fig A), however we observed significant increase in the number of hepatoblasts in L2 conditions (806 cells) compared to L0 and L1 (490 and 477 respectively). There were 5 different clusters for hepatocytes indicating hepatocyte heterogeneity. Differential gene expression analysis identified around 40 genes among the L1 vs L0 and L2 vs L0. The most significantly decreased gene is CYP2C19 while the most drastically increased genes were solute carrier gene (SLC) such as SLC1A1 and SLC22A4 and their expression are shown in dimension reduction plot (UMAP) (Fig B)
*Conclusions: Our findings will enhance the understanding of specific cellular response to the cold preservation and IRI at single cell resolution. Genes and pathways identified at single cell resolution will guide us to develop better methods for tissue preservation and treatment for liver IRI.
To cite this abstract in AMA style:
Naik S, Dogan M, Watkins C, Bajwa A, Eymard C, Eason J, Kuscu C, Kuscu C. Characterization of Hepatocytes During Cold Static Preservation at Single Cell Resolution [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/characterization-of-hepatocytes-during-cold-static-preservation-at-single-cell-resolution/. Accessed February 20, 2026.« Back to 2022 American Transplant Congress