Beyond Microarrays: Insights from Ncounter Transcript Analysis of Routine Archival Kidney Allograft Biopsies
Massachusetts General Hospital, Boston, MA
Meeting: 2021 American Transplant Congress
Abstract number: 318
Keywords: Biopsy, Kidney transplantation, Rejection
Topic: Clinical Science » Kidney » Kidney Chronic Antibody Mediated Rejection
Session Information
Session Name: Kidney Antibody Mediated Rejection
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 8, 2021
Session Time: 4:30pm-5:30pm
 Presentation Time: 4:35pm-4:40pm
Presentation Time: 4:35pm-4:40pm
Location: Virtual
*Purpose: Microarray mRNA analysis of renal allograft biopsies has discovered important insights, but has limitations as a practical clinical test. Here we evaluate a new technique that quantifies transcripts from routine formalin fixed paraffin embedded (FFPE) tissue.
*Methods: Gene expression was measured from 140 renal allograft biopsy samples (2003-2020): 45 chronic, active antibody mediated rejection (CAMR), 38 T cell mediated rejection (TCMR), 26 without evidence of rejection (NER), 19 borderline/suspicious (BS), 4 C4d+ NER, 8 living donor and 8 control native biopsies. RNA was extracted from 3 FFPE 20 μm sections and hybridized with the Banff Human Organ Transplant (B-HOT) 758 gene panel (NanoString) and quantitated (nCounter Max). Data were analyzed using the NanoString software with custom pathways and results correlated with Banff scores from the same biopsy core and outcome at 3 years.
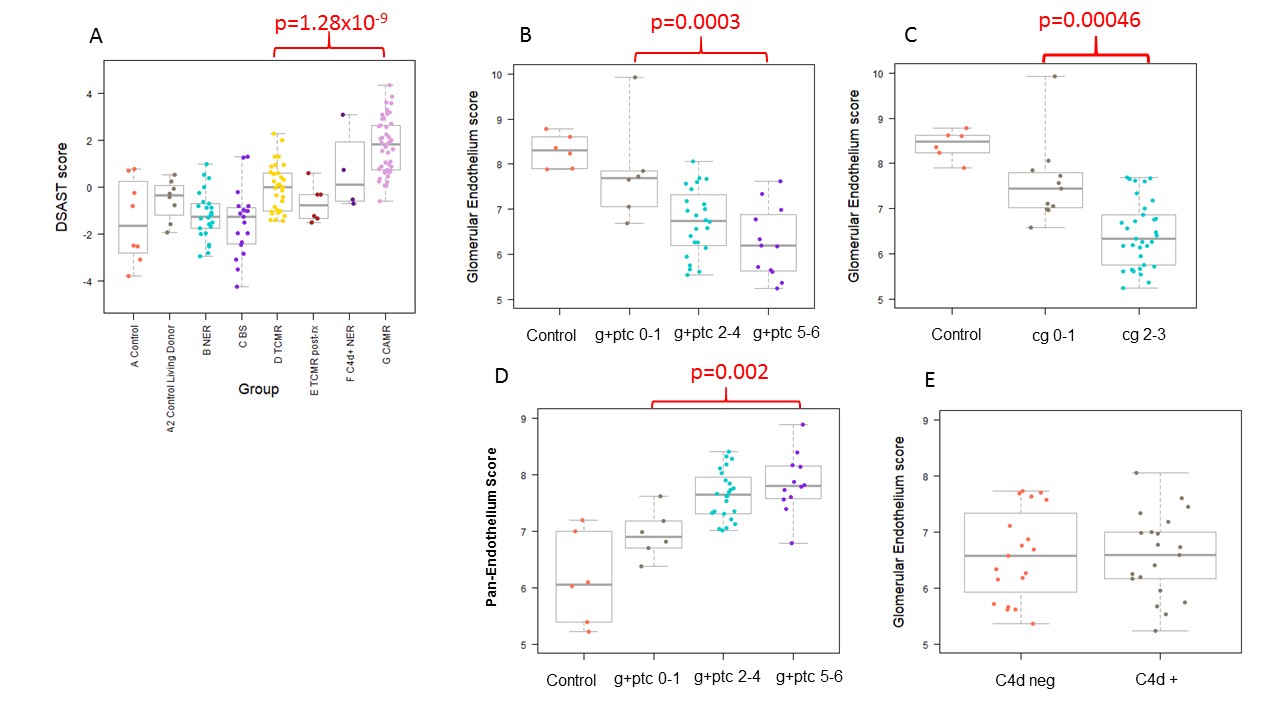
*Results: Many gene sets discriminated CAMR from TCMR. The best were DSAST (Hidalgo 2011, p=1.28×10-9 Fig 1A, AUC 87.5%) and TCMR (Venner 2014; p=1.3×10-8;AUC 84.9%), both derived from microarray studies, and CAMR NHP (Smith 2018; p=10-6). TCMR pathway scores were reduced in responsive post-treatment biopsies. C4d+ CAMR had higher levels of AMR, B and T cell pathways than C4d- CAMR. Selective glomerular injury could be detected in CAMR. Biopsies with higher microvascular inflammation (g+ptc, Fig 1B) and transplant glomerulopathy (cg, Figure 1C) had a progressive reduction of glomerular endothelial transcripts (EHD3, SOST) and podocyte specific transcripts (NPHS1, NPHS2), but these were not affected by C4d deposition. In contrast, pan-endothelial transcripts (e.g., CAV1, CDH5, PECAM1) were increased (Fig 1D). Graft loss at 3 years after TCMR/BS was rare, but those that failed showed higher endothelial scores due to occult AMR. Graft loss at 3 years in CAMR was associated with higher damage pathway scores but was not associated with increased AMR pathways.
*Conclusions: Robust transcript data can be obtained from routine archival FFPE clinical biopsies using NanoString technology and the B-HOT panel. Several pathway scores distinguish CAMR and TCMR with reasonable ROC performance. New findings suggest microvascular inflammation, independent of complement activation, causes glomerular injury in CAMR. Damage pathway scores rather than AMR or TCMR pathways predicted CAMR outcomes, but AMR pathways were predictive of rare graft failure in TCMR/BS biopsies.
To cite this abstract in AMA style:
Rosales I, Tomaszewski K, Milagros A, Kawai T, Smith R, Colvin R. Beyond Microarrays: Insights from Ncounter Transcript Analysis of Routine Archival Kidney Allograft Biopsies [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/beyond-microarrays-insights-from-ncounter-transcript-analysis-of-routine-archival-kidney-allograft-biopsies/. Accessed March 10, 2026.« Back to 2021 American Transplant Congress