A Universal Strategy to Characterize Allograft Infiltrating Cells by Bioinformatics Deconvolution of Next Generation Sequencing Data.
1Weill Cornell Medicine-Qatar, Doha, Qatar
2Weill Cornell Medicine, New York
Meeting: 2017 American Transplant Congress
Abstract number: 88
Session Information
Session Name: Concurrent Session: Predicting Tolerance and Rejection
Session Type: Concurrent Session
Date: Sunday, April 30, 2017
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:42pm-3:54pm
Presentation Time: 3:42pm-3:54pm
Location: E351
RNA sequencing characterizes mRNA transcriptome at an unprecedented level of precision. Because organ transplantation is a unique situation of mRNAs encoded by two (donor and recipient) genomes sequenced simultaneously, we investigated the novel hypothesis that the Het/Hom ratio (ratio of heterozygous genotypes to non-reference allele homozygous genotypes), currently used to identify contamination in DNA-Seq data, is informative of allograft-infiltrating cells.
We isolated total RNA from 42 human kidney allograft biopsies from 42 recipients; 37 for-cause biopsies (acute cellular rejection-ACR=7; antibody rejection-AMR=8; plasma cell-rich acute rejection-PCRR=7, transplant glomerulopathy-TG=9 and interstitial fibrosis/tubular atrophy-IFTA=6) and 5 surveillance biopsies reported as normal (Normal=5).
We used Illumina HiSeq to generate RNA-Seq data and GATK best practice protocol for RNA-Seq variant calling using iGenome Ensembl gene annotation. We calculated Het/Hom ratios using an in-house python script. We used ESTIMATE algorithm, where a score of 1=no admixture of recipient 's graft infiltrating cells (GICs) and donor kidney parenchymal cells in the biopsy.
ACR biopsies with GICs had the highest Het/Hom ratio (median 1.29) and the Normal biopsies without the GICs had the lowest ratio (median 0.898).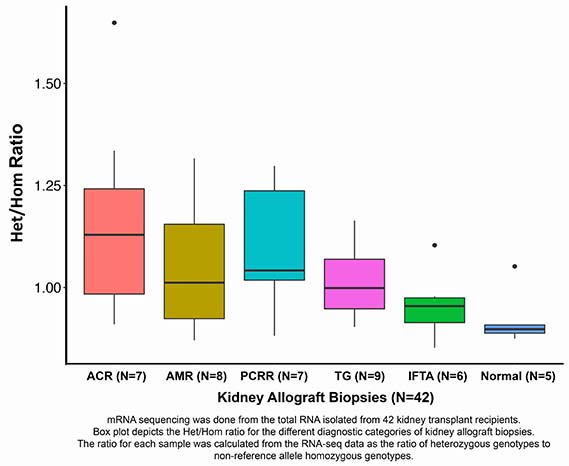 Het/Hom ratios were significantly associated with the Banff inflammation score (r=0.55, P<0.001) and the ESTIMATE purity score (r=0.77, P<0.0001).
Het/Hom ratios were significantly associated with the Banff inflammation score (r=0.55, P<0.001) and the ESTIMATE purity score (r=0.77, P<0.0001). Our demonstration that bioinformatics deconvolution of RNA-Seq data yields Het/Hom ratios reflective of recipient cells' infiltrating human kidney allografts, offers a universal approach to monitor cellular infiltration into any type organ grafts.
Our demonstration that bioinformatics deconvolution of RNA-Seq data yields Het/Hom ratios reflective of recipient cells' infiltrating human kidney allografts, offers a universal approach to monitor cellular infiltration into any type organ grafts.
CITATION INFORMATION: Thareja G, Yang H, Hayat S, Li C, Mueller F, Snopkowski C, Perry L, Magruder M, Lee J, Dadhania D, Suhre K, Suthanthiran M, Muthukumar T. A Universal Strategy to Characterize Allograft Infiltrating Cells by Bioinformatics Deconvolution of Next Generation Sequencing Data. Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Thareja G, Yang H, Hayat S, Li C, Mueller F, Snopkowski C, Perry L, Magruder M, Lee J, Dadhania D, Suhre K, Suthanthiran M, Muthukumar T. A Universal Strategy to Characterize Allograft Infiltrating Cells by Bioinformatics Deconvolution of Next Generation Sequencing Data. [abstract]. Am J Transplant. 2017; 17 (suppl 3). https://atcmeetingabstracts.com/abstract/a-universal-strategy-to-characterize-allograft-infiltrating-cells-by-bioinformatics-deconvolution-of-next-generation-sequencing-data/. Accessed March 3, 2026.« Back to 2017 American Transplant Congress
