Mining the Human Proteome for Monitoring Renal Transplant Injury.
1Surgery, University of California-San Francisco, San Francisco
2Pacific Northwest National Laboratory, Richland.
Meeting: 2016 American Transplant Congress
Abstract number: 206
Keywords: Kidney transplantation, Rejection, Renal injury
Session Information
Session Name: Concurrent Session: Kidney Immune Monitoring 2
Session Type: Concurrent Session
Date: Monday, June 13, 2016
Session Time: 2:30pm-4:00pm
 Presentation Time: 2:30pm-2:42pm
Presentation Time: 2:30pm-2:42pm
Location: Ballroom C
The human urinary proteome reflects systemic and inherent renal injury perturbations and can be harnessed to define biomarkers for different kidney transplant injury states. Through a NIH-funded study, we sought to identify and validate panels of urine proteins that could serve as non-invasive surrogate biomarkers for kidney transplant injuries.
396 unique urine samples were collected contemporaneously with an allograft biopsy from 396 unique kidney transplant recipients. Centralized, blinded histology on the graft phenotyped matched urine samples into categories of acute rejection (AR), chronic allograft nephropathy (CAN), BK virus nephritis (BKVN), and stable (STA). A strategy involving liquid chromatography–mass spectrometry (LC-MS) based shotgun proteomics using iTRAQ based discovery (n=108) to quantify pooled samples and global label-free LC-MS analyses of individual samples (n=137) and targeted validation with selected reaction monitoring (SRM; n=151) was applied to identify and validate minimal urine protein/peptide biomarkers to accurately segregate organ injury causation and pathology. The panel of peptides was selected with an algorithm called LASSO and its extension elastic net.
A total of 958 proteins were initially quantified by iTRAQ, 87% of which were also identified among 1574 urine proteins detected in label-free LC-MS validation. 103 urine proteins were significantly (p<0.05) perturbed in graft injury and enriched for humoral immunity, complement activation and lymphocyte trafficking. A set of 131 peptides corresponding to 78/103 proteins were measured by SRM and validated as significant in an independent sample cohort. A rigorous A minimal set of 35 peptides mapping to 33 proteins, were modeled to segregate different injury groups (AUC =93% for AR, 99% for CAN, 83% for BKVN) [Fig1].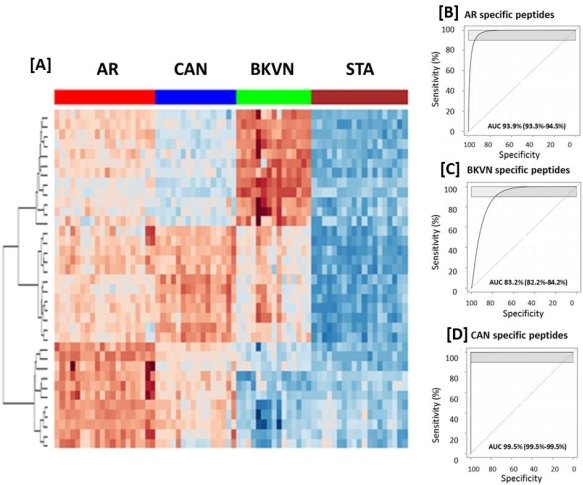
This translational research used biomarker discovery and validation used mass spectrometry based urinary proteomics and has identified urine protein fingerprints for non-invasive differentiation of kidney transplant injuries, thus opening the door for personalized immune risk assessment and therapy.
CITATION INFORMATION: Sigdel T, Gao Y, He J, Wang A, Nicora C, Smith D, Qian W, Camp D, Sarwal M. Mining the Human Proteome for Monitoring Renal Transplant Injury. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Sigdel T, Gao Y, He J, Wang A, Nicora C, Smith D, Qian W, Camp D, Sarwal M. Mining the Human Proteome for Monitoring Renal Transplant Injury. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/mining-the-human-proteome-for-monitoring-renal-transplant-injury/. Accessed February 18, 2026.« Back to 2016 American Transplant Congress
