Single Cell Rna-Sequencing of Urinary Cells and Defining the Immune Landscape of Rejection in Human Kidney Allografts
1Nephrology, NY Presbyterian- Weill Cornell Medical College, New York, NY, 2Physiology and Biophysics, Weill Cornell Medicine-Qatar, Doha, Qata
Meeting: 2021 American Transplant Congress
Abstract number: 305
Keywords: Kidney transplantation, Rejection, T cells, Transcription factors
Topic: Basic Science » Biomarker Discovery and Immune Modulation
Session Information
Session Name: Biomarkers and Cellular Therapies
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 8, 2021
Session Time: 4:30pm-5:30pm
 Presentation Time: 4:50pm-4:55pm
Presentation Time: 4:50pm-4:55pm
Location: Virtual
*Purpose: Transcriptome-based clustering of urinary cells in kidney transplant recipients may help identify cell-type specific injury and develop cell-type specific biomarkers for the noninvasive assessment of human kidney allografts.
*Methods: Single cell RNA-sequencing (scRNA-seq) was performed on urinary cells obtained at the time of allograft biopsy from kidney transplant recipients with biopsies classified as acute T cell mediated rejection [TCMR], chronic active antibody mediated rejection [ABMR], or normal/no rejection [Normal]. Droplet-based 10x Chromium platform (10x Genomics) was used to capture the individual urinary cells in emulsion, followed by cDNA synthesis, sequencing, and data analysis.
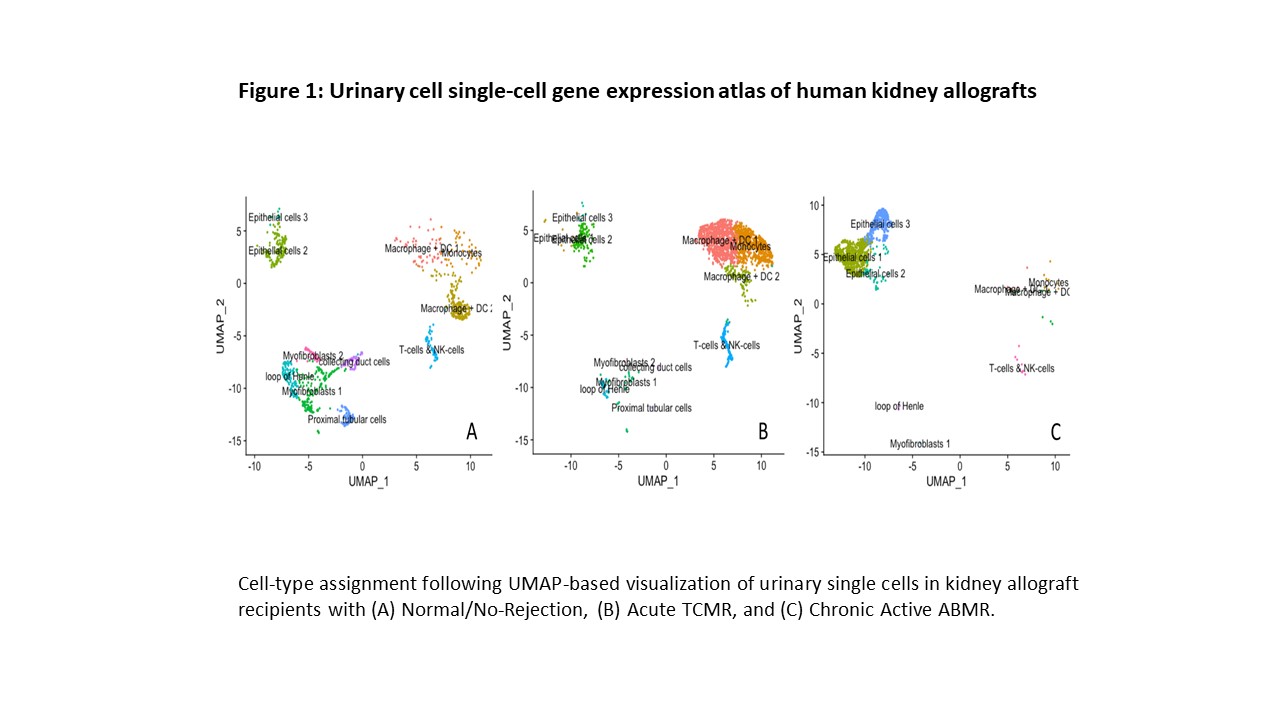
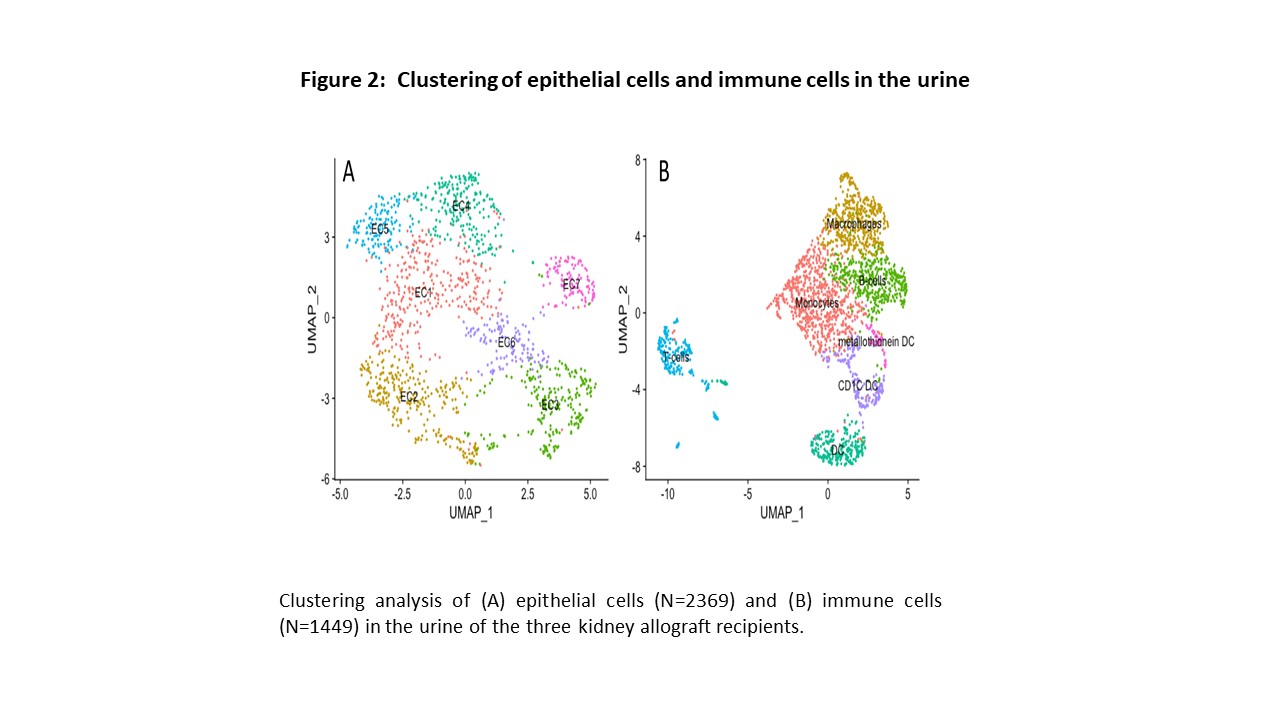
*Results: We obtained 3947 high quality cellular transcriptomes from the urine samples matched to TCMR biopsy, ABMR biopsy, or Normal biopsy. Cell-type assignment following uniform manifold approximation and projection (UMAP)-based visualization of urinary single cells from the patient with Normal/No-Rejection biopsy (Panel A), acute TCMR biopsy (Panel B), or ABMR biopsy (Panel C) are shown in Figure 1. Urine samples matched to the TCMR biopsy was exemplified by increased macrophages, dendritic cells, T cells, and NK cells whereas renal tubular epithelial cells were dominant in urine matched to normal biopsy. Further resolution of immune cells revealed sub-clusters of dendritic cells (Figure 2).
*Conclusions: We have demonstrated the feasibility of transcriptome analysis of individual cells in urine samples matched to human kidney allograft biopsies. To our knowledge, this is the first report of urinary cell scRNA-seq in kidney transplant recipients. Our study has deciphered the complex cellular landscape of kidney allograft rejection and provides opportunity to interrogate molecular events at a hitherto unattained level of resolution.
To cite this abstract in AMA style:
Muthukumar T, Yang H, Belkadi A, Thareja G, Li C, Snopkowski C, Chen K, Salinas T, Lubetzky M, Lee J, Dadhania D, Suhre K, Suthanthiran M. Single Cell Rna-Sequencing of Urinary Cells and Defining the Immune Landscape of Rejection in Human Kidney Allografts [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/single-cell-rna-sequencing-of-urinary-cells-and-defining-the-immune-landscape-of-rejection-in-human-kidney-allografts/. Accessed February 7, 2026.« Back to 2021 American Transplant Congress