Peripheral Blood RNA Sequencing Unravels a Differential Role of Coding and Non-Coding Genes by Types of Kidney Allograft Rejection
1University of California San Francisco, San Francisco, CA, 2Laboratory of Experimental Nephrology and Transplantation, Barcelona, Spain
Meeting: 2020 American Transplant Congress
Abstract number: 89
Keywords: Antibodies, Kidney, Rejection, T cell activation
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes I
Session Type: Oral Abstract Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:45pm
 Presentation Time: 3:15pm-3:27pm
Presentation Time: 3:15pm-3:27pm
Location: Virtual
*Purpose: Peripheral blood molecular patterns characterizing the different effector immune pathways driving kidney allograft rejection remain to be fully completely elucidated, to more fully type the gene signatures that may differentiate cellular and humoral graft rejection. We hypothesized that high resolution transcriptome analysis using RNA sequencing (RNAseq) in biopsy-matched peripheral blood (PB) samples of kidney transplant patients with different types of allograft rejection, would enable identifying unique protein-coding and non-coding genes segregating main transplant rejection types, unlike previous studies using microarray technology alone.
*Methods: To this end, we evaluated 100 biopsy-paired PB Samples from three distinct kidney transplant cohorts, with stable[STA], antibody-mediated rejection [AMR] and T-cell mediated rejection [TCMR], by RNAseq. Advanced machine learning tools were used to perform three way differential gene expression analysis on the first cohort to identify coding and on-coding genes with greatest specificity for AMR diagnosis.
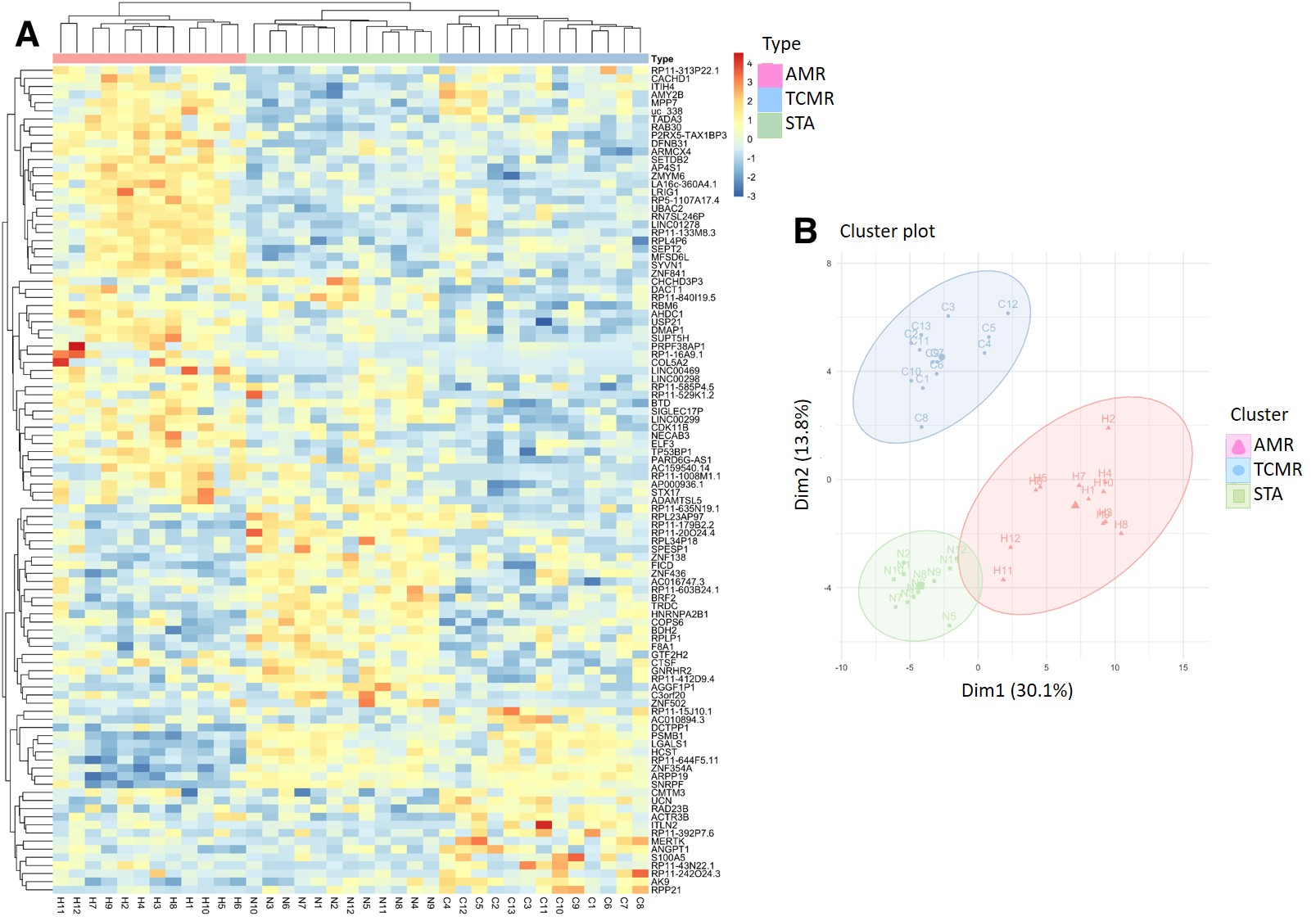
*Results: 102 genes (63 coding and 39 non-coding genes) associated with AMR (54up-regulated), TCMR (23 up-regulated) and STA (25up-regulated) perfectly clustered with each rejection phenotype and highly correlated with main fundamental histological lesions defining each rejection type (rho = 0.91). Functional analysis of the genes associated with AMR phenotype found enrichment in regulation of endoplasmic reticulum stress, adaptive immunity, and immunoglobulin class-switching. Subsequently, independent validation of target mRNAs was conducted by Fluidigm in two additional kidney transplant cohorts. The SIGLEC17P pseudogene and 9 SIGLEC17P-related coding genes, an NK cell gene, were highly regulated in AMR, clearly differentiating TCMR and STA samples.
*Conclusions: In conclusion, PB RNASeq identifies a critical role for PB non-coding genes in AMR, highlighting a reason why prior studies on coding genes alone have been unable to differentiate AMR from TCMR. Targeted SIGLEC17P gene family amplification is a valuable biomarker for perturbations of NK biology in AMR, and rapid amplification based assessment of these genes open the door for customized immunosuppression therapy for graft rejection.
To cite this abstract in AMA style:
Sur S, Pineda S, Sigdel T, Sirota M, Bestard O, Sarwal M. Peripheral Blood RNA Sequencing Unravels a Differential Role of Coding and Non-Coding Genes by Types of Kidney Allograft Rejection [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/peripheral-blood-rna-sequencing-unravels-a-differential-role-of-coding-and-non-coding-genes-by-types-of-kidney-allograft-rejection/. Accessed January 7, 2026.« Back to 2020 American Transplant Congress