Replication of Diagnostic Biomarkers Using Orthogonal State-of-the-Art Technologies
WCM, New York, NY
Meeting: 2019 American Transplant Congress
Abstract number: A22
Keywords: Biopsy, Kidney transplantation, Renal function
Session Information
Session Name: Poster Session A: Acute Rejection
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: The failure to reproduce scientific findings (McNutt M. Science 2014;343:229, Baker M. Nature 2016;53:452) has brought to the forefront the need for replication studies prior to their clinical application. We therefore investigated whether the previously discovered urinary cell biomolecular markers of kidney allograft rejection could be replicated in an independent cohort of kidney graft recipients. We applied two orthogonal technologies to investigate biomarker data reproducibility: RNA-sequencing (RNA-Seq), currently the best tool for unbiased and precise characterization of mRNA transcriptome, and pre-amplification enhanced real time quantitative PCR (RT-qPCR) assays for absolute quantification of mRNAs.
*Methods: We performed RNA-Seq of 46 biopsy-matched urinary cell specimens from 46 kidney recipients; 22 acute T cell mediated rejection (ACR) and 24 no rejection (NR). We analyzed the urine RNA-Seq data to determine whether urinary cell abundance of CD3E, IP-10 (CXCL10), granzyme B (GZMB), perforin (PRF1), and CD103 (ITGAE) are diagnostic of ACR as reported. We used RT-qPCR assays for absolute quantification of these mRNAs and investigated whether the previously reported associations are reproducible.
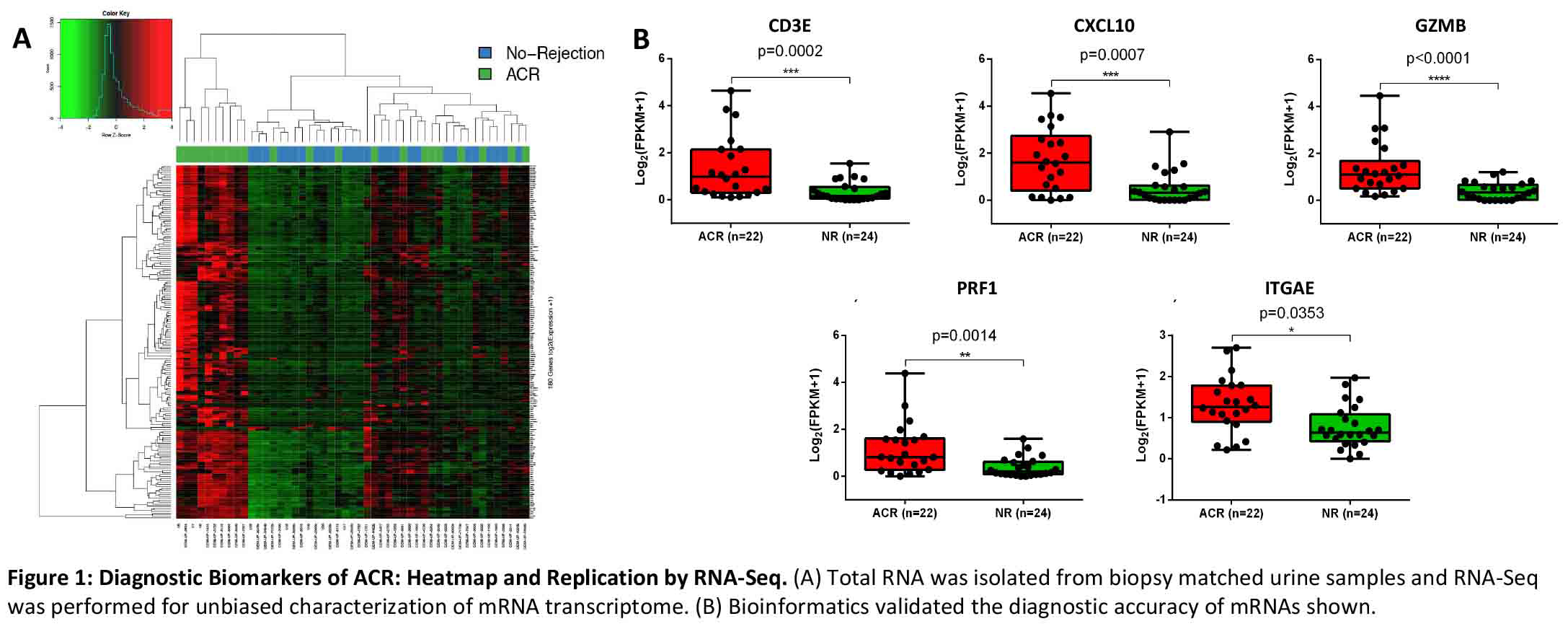
*Results: In complete accordance with previously published data, the abundance of transcripts for CD3E, CXCL10, GZMB, PRF1 and ITGAE, ascertained by RNA-Seq were significantly higher in urine matched to ACR biopsies compared to urine matched to NR biopsies (Figure 1).
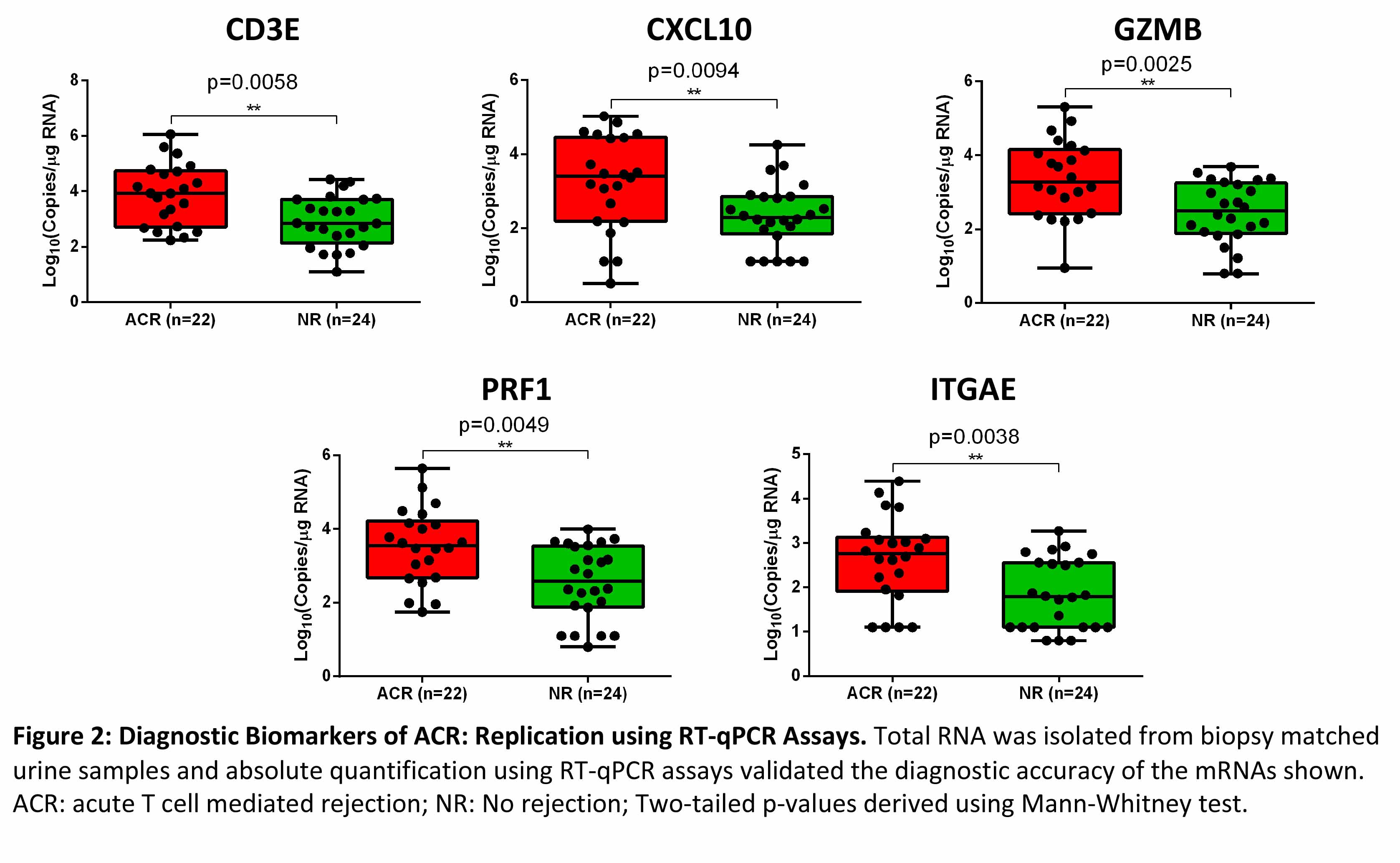
Absolute quantification of mRNA copies using RT-qPCR assays demonstrated that copies of mRNA CD3E, CXCL10, GZMB, PRF1 and ITGAE were significantly higher in urine matched to ACR biopsies compared to urine matched to NR biopsies (Figure 2).
*Conclusions: We have replicated, with the use of state-of-the-art and orthogonal molecular tools, that urinary cell levels of mRNA for CD3E, CXCL10, GZMB, PRF1, and ITGAE are diagnostic of ACR in human kidney allografts. Altogether, the demonstrated reproducibility represents a significant step towards clinical application of urinary cell mRNAs for the noninvasive diagnosis of ACR in human kidney allografts.
To cite this abstract in AMA style:
Cassidy M, Dooley B, Verma A, Yang H, Muthukumar T, Ding R, Li C, Snopkowski C, Botticelli B, Albakry S, Edusei E, Lubetzky M, Dadhania D, Lee J, Elemento O, Suthanthiran M. Replication of Diagnostic Biomarkers Using Orthogonal State-of-the-Art Technologies [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/replication-of-diagnostic-biomarkers-using-orthogonal-state-of-the-art-technologies/. Accessed March 3, 2026.« Back to 2019 American Transplant Congress