Histogenomics: Molecular Profiling of Kidney Biopsies Predict Delayed Graft Function
1NJ Sharing Network, New Providence, NJ
2Rutgers Robert Wood Johnson Medical School, New Brunswick, NJ.
Meeting: 2018 American Transplant Congress
Abstract number: D104
Keywords: Gene expression, Kidney, Renal function
Session Information
Session Name: Poster Session D: Kidney Complications: Late Graft Failure
Session Type: Poster Session
Date: Tuesday, June 5, 2018
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall 4EF
Introduction
Histologic assessments of donor renal biopsies for percentage global glomerulosclerosis, interstitial fibrosis and arterial sclerosis can predict renal graft failure post-transplant.
The morphologic evaluation of renal biopsies does not provide information on the functional status of the kidney or the pathophysiological causes that lead to delayed graft function (DGF). We hypothesized that molecular profiling of kidney biopsies may have an additive effect in predicting post-transplant allograft function.
Methods
Total RNA that was extracted from the renal biopsies and was reverse transcribed to cDNA and overlaid onto 96-well PCR plates containing customized gene-specific PCR primer pairs for genes selected based on broad review of the literature on DGF. The plates were then processed using quantitative Real Time PCR and results reported as fold-changes in gene expression and compared between kidneys with primary versus delayed graft function.
Results
We compared 16 biopsies that went on to show DGF (defined by return to dialysis within first week) with 16 biopsies from kidneys that displayed primary function post-transplant. We observed increased expression of 4 genes that were differentially regulated in DGF cases. 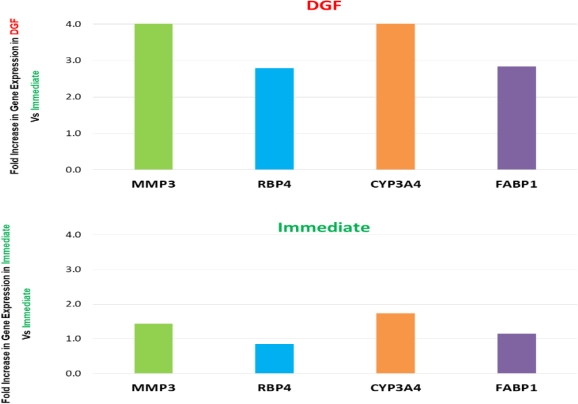 These genes represented different gene ontology categories such as inflammation and immune response and cell growth/metabolism.
These genes represented different gene ontology categories such as inflammation and immune response and cell growth/metabolism.
| Inflamation and immune response | MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| General metabolism | RBP4 | retinol-binding protein 4, plasma |
| Lipid metabolism | CYP3A4 | cytochrome P450, subfamily IIIA (niphedipine oxidase), polypeptide 4 |
| Lipid metabolism | FABP1 | fatty acid binding protein 1, liver |
These same genes were minimally expressed in cases that showed primary function post-transplant. Our findings suggest that there is a disruption of the various intra-cellular functions in deceased donor kidneys that is highly associated with DGF post-transplant.
Conclusion
We have identified a gene fingerprint in transplant kidney biopsies that could be predictive of DGF.
CITATION INFORMATION: Rao P., Deo D., Fyfe-Kirschner B., Siderits R. Histogenomics: Molecular Profiling of Kidney Biopsies Predict Delayed Graft Function Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Rao P, Deo D, Fyfe-Kirschner B, Siderits R. Histogenomics: Molecular Profiling of Kidney Biopsies Predict Delayed Graft Function [abstract]. https://atcmeetingabstracts.com/abstract/histogenomics-molecular-profiling-of-kidney-biopsies-predict-delayed-graft-function/. Accessed February 10, 2026.« Back to 2018 American Transplant Congress
