Genome-Wide Association Study Assessing Risk of Rejection in Pediatric Transplant Recipients
1SickKids Hospital, Toronto, ON, Canada
2Winnipeg Children's Hospital, Winnipeg, MB, Canada
3BC Children's Hospital, Vancouver, BC, Canada
4Montreal Children's Hospital, Montreal, QC, Canada
5Alberta Children's Hospital, Calgary, AB, Canada
6CHU Sainte Justines, Montreal, QC, Canada
7Stollery Children's Hospital, Edmonton, AB, Canada.
Meeting: 2018 American Transplant Congress
Abstract number: A444
Keywords: Genomic markers, Pediatric, Rejection
Session Information
Session Name: Poster Session A: Tolerance: Clinical Studies
Session Type: Poster Session
Date: Saturday, June 2, 2018
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall 4EF
Background: Rejection risk varies after solid organ transplantation (SOT). We explored genomic variants associated with susceptibility to graft rejection.
Methods: SOTs enrolled in a multicenter prospective cohort study were genotyped using the Axiom Transplant Array. Patients experiencing rejection 1-year post transplant were compared to non-rejectors. GWAS was used to assess the association of SNP with rejection risk using Cox proportional hazard and logistic regression models. 490,311 variants with a minor allele frequency of >0.01 in Hardy-Weinberg equilibrium were included. Mutation burden was performed on significant SNPs. Gene set enrichment analysis (GSEA) was used to identify gene signatures. Co-variates included were organ type, sex, age, race, ABO compatibility and induction medication use.
Results: Of 486 participants (140 renal, 153 heart, 181 liver, 12 lung), 42% experienced rejection, median time to rejection was 11.24 months. Males had a lower hazard ratio (HR) for rejection than females (HR=0.75, p<0.05), while heart and liver had a higher hazard for rejection compared to kidney recipients (p< 0.001). 20 SNPs were associated with rejection (HR range 1.64-7.95, p<10-5). 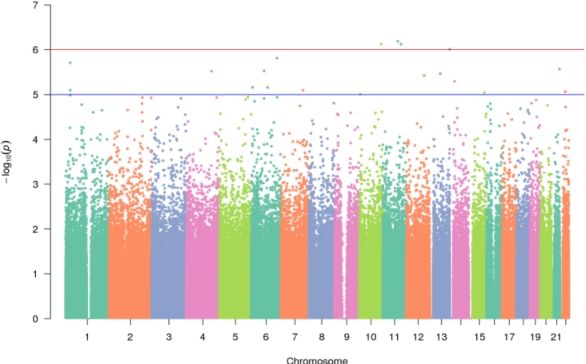 On CoxPH analysis, organ type, younger age and variant burden were associated with rejection with every additional SNP increasing risk by a HR of 1.38 (p <0.001). GSEA identified enrichment of variants in genes related to adaptive immune response, autoimmune disease, interferon signaling and regulation of autophagy.
On CoxPH analysis, organ type, younger age and variant burden were associated with rejection with every additional SNP increasing risk by a HR of 1.38 (p <0.001). GSEA identified enrichment of variants in genes related to adaptive immune response, autoimmune disease, interferon signaling and regulation of autophagy.
Conclusion: Variants in immunity associated genes are associated with rejection. Clinical and genetic factors can inform the development of individualized risk prediction models for rejection.
CITATION INFORMATION: Papaz T., Min S., Lee O., Blydt-Hansen T., Allen U., Avitzur Y., Birk P., Foster B., Grasemann H., Hamiwka L., Parekh R., Phan V., Urschel S., Mital S. Genome-Wide Association Study Assessing Risk of Rejection in Pediatric Transplant Recipients Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Papaz T, Min S, Lee O, Blydt-Hansen T, Allen U, Avitzur Y, Birk P, Foster B, Grasemann H, Hamiwka L, Parekh R, Phan V, Urschel S, Mital S. Genome-Wide Association Study Assessing Risk of Rejection in Pediatric Transplant Recipients [abstract]. https://atcmeetingabstracts.com/abstract/genome-wide-association-study-assessing-risk-of-rejection-in-pediatric-transplant-recipients/. Accessed January 29, 2026.« Back to 2018 American Transplant Congress
