Early Experience Using Donor-Derived Cell-Free DNA for Detection of Rejection in Kidney Transplantation
Medicine/Nephrology, Cedars Sinai Medical Center, Los Angeles, CA.
Meeting: 2018 American Transplant Congress
Abstract number: A110
Keywords: Kidney transplantation, Rejection
Session Information
Session Name: Poster Session A: Kidney Acute Antibody Mediated Rejection
Session Type: Poster Session
Date: Saturday, June 2, 2018
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall 4EF
Introduction:
Donor-derived cell free DNA (ddDNA; Allosure, CareDX) has been validated as a non-invasive test that can discriminate rejection from non-rejection in kidney transplantation, but has not been externally validated. We describe our early experience using ddDNA and report performance characteristics of ddDNA for discrimination of rejection.
Methods:
We measured ddDNA on 25 kidney transplant recipients, 12 of whom underwent confirmatory allograft biopsy. Receiver operating characteristic curves (ROC) were generated for any rejection (AR; antibody- or cell-mediated, CMR) and antibody-mediated rejection (ABMR) alone. The area under the ROC curve (AUC) and associated positive (PPV) and negative predictive values (NPV) were calculated.
Results:
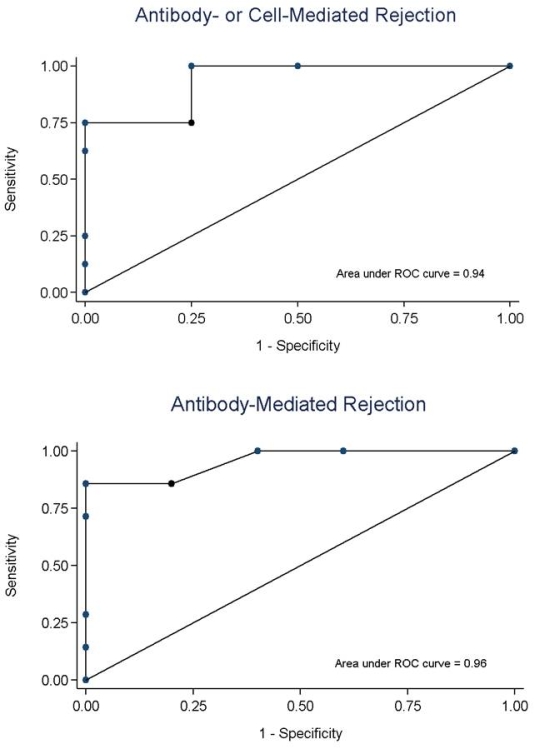
Blood for ddDNA was taken at a median 1125 days post-transplant (IQR: 338, 2844). The median GFR was 48.2 ml/min/1.73 m2 (IQR: 40.2-65.8). Donor-specific antibodies were present in 11/25 (44%). The median ddDNA was 1.0% (IQR: 0.3-1.5). Twelve patients had ddDNA below or near the lower limit of detection (≤0.29%); two were biopsied and did not have rejection (ddDNA: undetectable and 0.28%). Twelve patients underwent biopsy (median ddDNA 1.7%, IQR: 1.3-2.5), of whom 7 had ABMR and 1 had CMR. Two with ddDNA≥1.0% had no rejection on biopsy (ddDNA 1.2%: negative; ddDNA 1.5%: C3 glomerulopathy). Discrimination for AR (AUC 93.8%, 95% CI: 79.1-100%) and ABMR (AUC 95.7%, 95% CI: 85.6-100%) was high (fig 1). A ddDNA threshold ≥1.0% for AR yielded a PPV of 80.0% and NPV 100%; increasing the threshold to ≥1.4% improved the PPV to 88.9% while maintaining NPV 100%. For ABMR, the optimal ddDNA threshold was ≥1.9% (PPV: 100%; NPV: 83.3%).
Conclusions:
Test performance of ddDNA was similar to previously reported internal validation data. Although the number assessed was small, our preliminary experience reinforces that ddDNA may be useful for discrimination of AR and ABMR from non-rejection states. Detection of high ddDNA in a patient with C3 glomerulopathy argues for further studies to evaluate the impact of glomerulonephritis on ddDNA test performance.
CITATION INFORMATION: Huang E., Vo A., Choi J., Peng A., Sethi S., Najjar R., Jordan S. Early Experience Using Donor-Derived Cell-Free DNA for Detection of Rejection in Kidney Transplantation Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Huang E, Vo A, Choi J, Peng A, Sethi S, Najjar R, Jordan S. Early Experience Using Donor-Derived Cell-Free DNA for Detection of Rejection in Kidney Transplantation [abstract]. https://atcmeetingabstracts.com/abstract/early-experience-using-donor-derived-cell-free-dna-for-detection-of-rejection-in-kidney-transplantation/. Accessed February 21, 2026.« Back to 2018 American Transplant Congress