Validating Molecular Microscope Readings and Estimating Agreement with Histology
ATAGC, Edmonton, Canada.
Meeting: 2018 American Transplant Congress
Abstract number: 455
Keywords: Biopsy, Kidney, Kidney transplantation
Session Information
Session Name: Concurrent Session: Kidney: Acute Cellular Rejection
Session Type: Concurrent Session
Date: Tuesday, June 5, 2018
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:06pm-3:18pm
Presentation Time: 3:06pm-3:18pm
Location: Room 6A
We previously described a microarray diagnostic system (MMDx) for rejection in 1208 kidney transplant biopsies (J. Reeve et al. JCI Insight 2 (12), 2017). The system assigns molecular phenotype using binary classifiers, and estimates rejection classes using archetypal clustering. The present study validated the system in a set of 537 new biopsies and studied the relationship to histology (available in 227 of the 537). Histology diagnoses in validation set 537 were similar to set 1208.
Biopsies distributed in PCA by rejection molecular phenotype: PC1=rejection; PC2 =ABMR vs. TCMR; PC3=ABMR stage (early-stage; fully-developed; late-stage) (Fig.1: PC2 vs 1, left panels; PC2 vs 3, right panels). Set 1208 is at the top (ABCD), and set 537 at the bottom (figure 1EFGH). Biopsies are colored by histology class (AB and EF) and molecular class (CD and GH). Relationships between molecular phenotype (position) and histology diagnoses (color) in set 537 were similar to those in set 1208, including separation of ABMR stage in PC3 (Fig. 1H). Thus MMDx performed similarly in the validation set.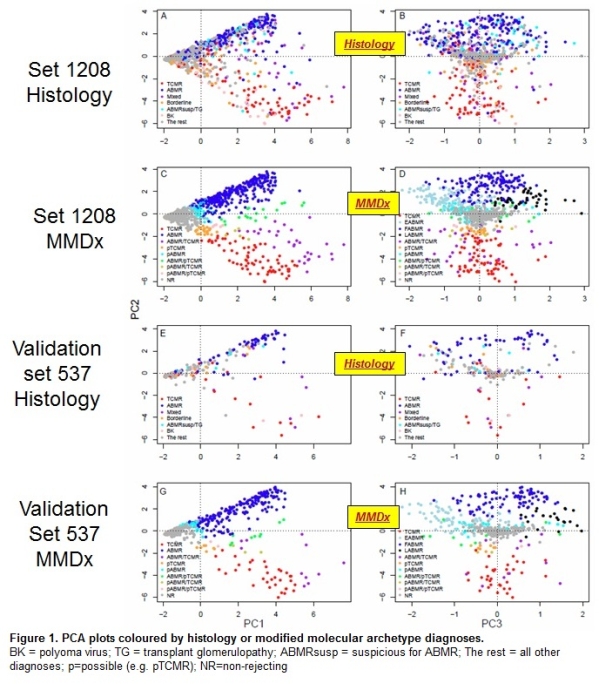
We used receiver-operator characteristic (ROC) curves to compare the ability of archetype analysis (AA) categories and gene set scores to predict histologic diagnoses (test set results) (Fig. 2A). AUCs were calculated for ABMR (Fig. 2A) or TCMR (Fig. 2B) in 1208, and in 227 biopsies in set 537 with histology diagnoses (Fig. 2 C,D). AA and binary classifiers performed similarly. Gene sets and single genes often performed less well than the machine learning methods (AA and binary classifiers), particularly in ABMR (Fig. 2A,C). The best AUC was for ABMR 0.86 and for TCMR 0.88. 
The results showed that the MMDx system created in 1208 biopsies performs similarly in validation set 537, and that machine learning classifiers rather than gene sets are necessary to optimize molecular diagnosis of rejection.
CITATION INFORMATION: Halloran P., Reeve J., The INTERCOMEX Study Group Validating Molecular Microscope Readings and Estimating Agreement with Histology Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Halloran P, Reeve J, Group TheINTERCOMEXStudy. Validating Molecular Microscope Readings and Estimating Agreement with Histology [abstract]. https://atcmeetingabstracts.com/abstract/validating-molecular-microscope-readings-and-estimating-agreement-with-histology/. Accessed January 31, 2026.« Back to 2018 American Transplant Congress
