Predicting Graft Failure in Kidney Transplant Patients: A Combined Clinical/Molecular Approach to Analyzing Biopsies
University of Alberta, Edmonton, AB, Canada
Hospital de la Vall d'Hebron, Barcelona, Spain
Manchester Royal Infirmary, Manchester, United Kingdom
Hannover Medical School, Hannover, Germany
University of Maryland School of Medicine, Baltimore, MD
University of Minnesota, Minneapolis, MN
Medicine, University of Alberta, Edmonton, AB, Canada
Meeting: 2013 American Transplant Congress
Abstract number: 258
We combined an existing dataset with new data from a multi-centre international collaboration (INTERCOM), to predict graft loss in a patient population undergoing kidney transplant biopsies for cause. We used a machine learning method – Random Survival Forests – combining histologic, clinical, and microarray data from 562 patients (1 biopsy per patient). Data were split into a training and test set, each with 55 failures in 281 patients. Random forests have a number of useful characteristics, including the ability to combine different data types and model interactions while minimizing problems associated with low sample size: number of variables ratio. In addition, a well established measure of variable importance is available for comparing the predictors.
Inputs included Banff lesion scores (t, i, g, ct, ci, cg, ptc), C4d staining, proteinuria (Prot), donor and recipient age (Dage, Rage), DSA, time of biopsy post-transplant (TxBx), and diagnosis of C4d+ or C4d- ABMR. In addition, three previously published molecular scores, IRRAT (injury response and repair transcripts), RS (molecular risk score) and ABMR score (probability of ABMR) were used.
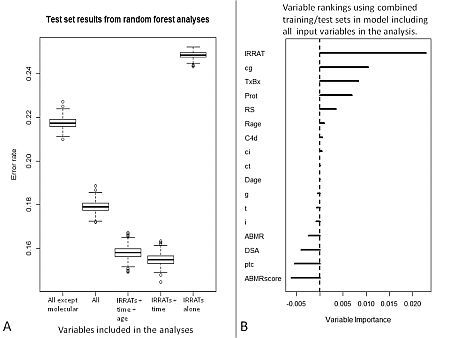
Fig. 1A shows the error rates (1 – Concordance Index) in the test set for models built using the training set. Boxplots are based on 1000 random forest iterations. Inclusion of molecular variables increased accuracy (All vs All except molecular), but the best model used only IRRATs and TxBx. IRRATs alone are poor predictors because injury is also high in early AKI and in TCMR patients whose kidneys rarely fail.
Figure 1B shows the importance of the inputs using all 562 patients (to obtain a better estimate of importance). Scores near 0.0 indicate non-informativeness. From the variables examined, the IRRATs are the best predictor of graft failure.
To cite this abstract in AMA style:
Reeve J, Sellares J, Freitas Dde, Einecke G, Bromberg J, Matas A, Halloran P. Predicting Graft Failure in Kidney Transplant Patients: A Combined Clinical/Molecular Approach to Analyzing Biopsies [abstract]. Am J Transplant. 2013; 13 (suppl 5). https://atcmeetingabstracts.com/abstract/predicting-graft-failure-in-kidney-transplant-patients-a-combined-clinicalmolecular-approach-to-analyzing-biopsies/. Accessed March 9, 2026.« Back to 2013 American Transplant Congress
