Immunohistochemical and Molecular Detection of NK Cells in Kidney Allografts with Antibody Mediated Rejection
Pathology, University of Michigan, Ann Arbor, MI
Meeting: 2020 American Transplant Congress
Abstract number: D-271
Keywords: CD3, Kidney transplantation, Natural killer cells, Protocol biopsy
Session Information
Session Name: Poster Session D: Biomarkers, Immune Assessment and Clinical Outcomes
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Molecular signatures specific to Natural Killer (NK) cells are closely associated with antibody mediated rejection (AMR) in transplant biopsies. We have developed an immunohistochemical (IHC) approach to detect NK cells in formalin fixed paraffin embedded (FFPE) tissue sections using CD16/Tbet dual staining to quantify NK cells within routine biopsy slides. Here we test this approach by comparing NK cells by paraffin immunohistochemistry against NK cell and other immune transcripts by Nanostring quantification from adjacent sections.
*Methods: 11 FFPE human kidney transplant biopsies were obtained from pathology archives, 3 surveillance biopsies from stable patients and 4 biopsies each of active AMR and chronic active AMR. 5 µm sections were stained for CD3, CD19, or CD16/Tbet, digitized, and quantified for T cells, B cells, and NK cells/mm2. Triplicate adjacent 10 µm FFPE sections were used for RNA extraction (Recoverall, Invitrogen), with overall RIN values ranging from 1.8-2.3. RNA was hybridized to the Immunology v2 probeset, and quantified using a Sprint Profiler and NSolver software (Nanostring). Probe counts were normalized to internal housekeeping transcripts and a water control on each chip. Using the surveillance biopsies as the reference, the unweighted average relative fold change of NK cell, T cell, and B cell-specific probe counts for each transcript set were compared to cell counts by IHC.
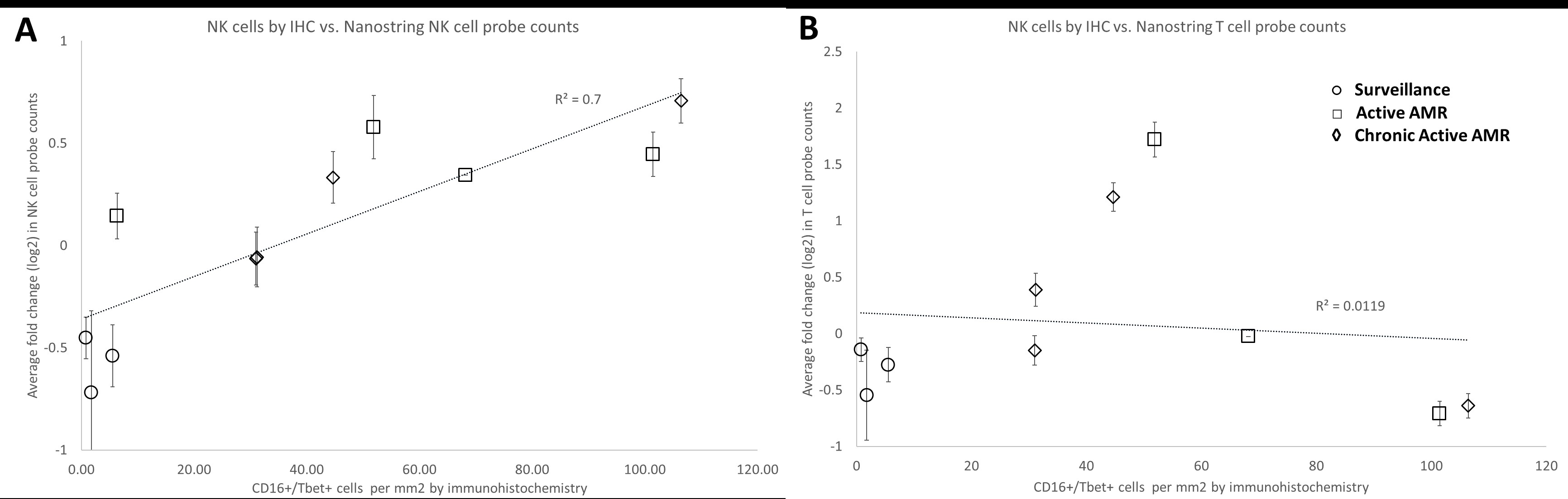
*Results: CD16+/Tbet+ NK cell densities ranged from 0.8-5.5 cells/mm2 tissue in surveillance biopsies and 6.3-105 cells/mm2 tissue in AMR biopsies. NK cell densities were directly correlated with Nanostring NK cell related probe counts averages (R2 = 0.7, Fig 1A), but showed no correlation with T cell (R2 = 0.0119, Fig 1B) or B cell specific probe counts (R2 < 0.01). Similarly, CD3+ cells/mm2 tissue and CD20+ cells/mm2 showed a direct correlation between average T cell (R2 = 0.69) and B cell (R2 = 0.62) probe counts respectively, but no correlation with NK cell-specific transcripts (R2 < 0.04).
*Conclusions: Dual CD16/Tbet staining effectively identifies NK cells in paraffin tissue in renal transplant biopsies with AMR. Detection of NK cells in FFPE by immunohistochemical or molecular methods may be a useful biomarker in AMR diagnosis.
| Cell type | Probes |
| NK cell | NCAM1, 7 KIR genes, NCR1, KLRAP1, KLRB1, KLRC1-4, KLRD1, KLRF1-2, KLRK1 |
| T cell | CD3D/E, CD4, CD8A/B, CD27, CD28, CD40LG, ICOS, CTLA4 |
| B cell | CD19, CD22, CD79A, CD79B, PAX5, TNFRSF17 |
To cite this abstract in AMA style:
Farkash E, Barnes J, Anand S. Immunohistochemical and Molecular Detection of NK Cells in Kidney Allografts with Antibody Mediated Rejection [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/immunohistochemical-and-molecular-detection-of-nk-cells-in-kidney-allografts-with-antibody-mediated-rejection/. Accessed March 9, 2026.« Back to 2020 American Transplant Congress