A “Multi-Omic” Analysis of Proteome and Metabolome for Acute Rejection and PVAN in Kidney Transplantation
Surgery, UCSF, San Francisco.
Meeting: 2018 American Transplant Congress
Abstract number: C183
Keywords: Kidney transplantation, Rejection
Session Information
Session Name: Poster Session C: Kidney: Polyoma
Session Type: Poster Session
Date: Monday, June 4, 2018
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall 4EF
Background: Most effort in identifying biomarkers have used gene expression analyses. Proteomics and metabolomics are useful to identify surrogate biomarkers in monitoring transplanted kidney and can help with our understanding of graft dysfunction and failure.
Method:308 urine samples (stable graft -STA, n=111, acute rejection -AR, n=106, chronic allograft nephropathy -CAN, n=69, were analyzed by GC–MS using a gas chromatograph interfaced to a time-of-fliight (TOF) mass spectrometer. The raw data was normalized against creatinine. Unpaired T-test, ANOVA and significant analysis of microarrays (SAM), were used to identify injury specific panel of metabolites. Previously reported proteomics data was used in “multi-omic” analysis. We used pathway enrichment and pathway topological analysis were performed.
Result: 266 metabolites were identified across the urine samples of which 146 metabolites were significantly altered (42 increased and 104 decreased) in AR compared to STA (P<0.05, 5.0% FDR). For BKVN versus STA comparison, 163 metabolites (90 increased and 73 decreased) (P<0.05, corresponding to a 5% FDR). 128 metabolites (91 increased and 37 decreased) were significantly altered in the IFTA urine samples compared to the AR samples, (P<0.05, 4.0% FDR). A “multi-omic” analysis that comprised of 404 AR-specific urine proteins and 146 AR-specific urine metabolites revealed enrichment of starch and sucrose metabolism(p=1.09E-05), galactose metabolism (p=2.16E-05), aminoacyl-tRNA biosynthesis (p=3.16E-05), glutathione metabolism (1.31E-03), and Glycine, serine and threonine metabolism (p=3.28E-03) at the time AR. Additional multi-omic analysis that comprised of 94 PVAN-specific urine proteins and 163 PVAN-specific metabolites revealed aminoacyl-tRNA biosynthesis (p=1.09E-05), beta-Alanine metabolism (p=2.16E-05) and pantothenate and CoA biosynthesis (p=3.16E-05) as most significant in PVAN (Figure 1).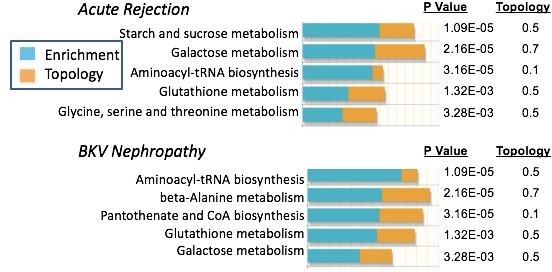
Conclusion:Integrative “multi-omic” analysis of urine proteome and metabolome provides some unique metabolic processes, with a remarkable overlap of many energetic pathways in kidney pathology with AR and PVAN, suggesting common tissue injury pathways triggered by both allo and innate immunity.
CITATION INFORMATION: Sigdel T., Yang J., Sarwal M. A “Multi-Omic” Analysis of Proteome and Metabolome for Acute Rejection and PVAN in Kidney Transplantation Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Sigdel T, Yang J, Sarwal M. A “Multi-Omic” Analysis of Proteome and Metabolome for Acute Rejection and PVAN in Kidney Transplantation [abstract]. https://atcmeetingabstracts.com/abstract/a-multi-omic-analysis-of-proteome-and-metabolome-for-acute-rejection-and-pvan-in-kidney-transplantation/. Accessed February 20, 2026.« Back to 2018 American Transplant Congress
