Cell Free DNA Sequencing for Diagnosing Infections, Estimating Growth Rates, and Characterizing Antibiotic Resistomes in Urine Supernatants
1Weill Cornell Medicine, New York, NY
2Cornell University, Ithaca, NY.
Meeting: 2018 American Transplant Congress
Abstract number: 440
Keywords: Infection, Kidney transplantation
Session Information
Session Name: Concurrent Session: Kidney Infectious - Pot-Pourri
Session Type: Concurrent Session
Date: Tuesday, June 5, 2018
Session Time: 2:30pm-4:00pm
 Presentation Time: 2:54pm-3:06pm
Presentation Time: 2:54pm-3:06pm
Location: Room 608/609
INTRODUCTION. We examined the utility of cell-free DNA (cfDNA) sequencing of urine supernatants in kidney transplant recipients for diagnosing rare and common urinary tract infections (UTI), estimating bacterial growth rates, and antibiotic resistant genes.
METHODS. We collected 28 urine specimens from 18 kidney transplant recipients with bacterial UTI, 6 urine specimens from 2 kidney transplant recipients with parvovirus infection, and 2 urine specimens from 1 kidney transplant recipient with adenovirus infection. Single-stranded library preparation and shotgun sequencing on the 36 urine supernatants was performed using Illumina Next Seq, 75 bp by 75 bp. Microbial identification was performed using the Grammy pipeline.
RESULTS. In 27 of the 28 bacterial UTIs, urinary cfDNA identified the pathogen confirmed by bacterial culture. Urinary cfDNA was able to detect H. influenza in a urine culture that was negative by conventional urine culture but was positive for H. influenza after the clinician requested advanced culturing techniques. In the cases of parovovirus and adenovirus infections, urinary cfDNA detected the pathogens in all cases at least 1 week prior to the clinical diagnosis made by the clinicians. Antibiotic resistomes were constructed based on urinary cfDNA in the bacterial UTI associated samples and correlated with antimicrobial susceptibility testing. 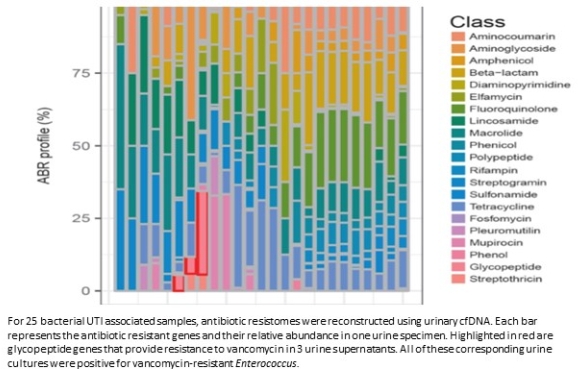
Growth rates of bacterial pathogens were estimated based on uneven cfDNA coverage over the origin of replication. 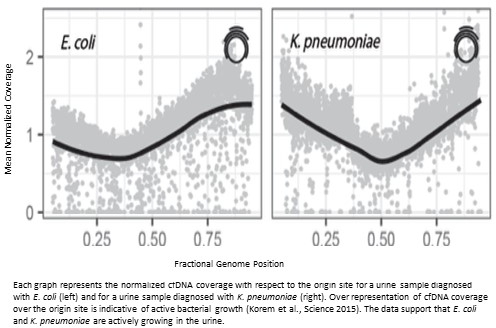
CONCLUSIONS. Cell free DNA sequencing in urine supernatants provides an all-inclusive method to anticipate and diagnose common and rare infections as well as inform antibiotic resistance patterns and bacterial growth rate.
CITATION INFORMATION: Lee J., Burnham P., Dadhania D., Heyang M., Chen F., Suthanthiran M., De Vlaminck I. Cell Free DNA Sequencing for Diagnosing Infections, Estimating Growth Rates, and Characterizing Antibiotic Resistomes in Urine Supernatants Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Lee J, Burnham P, Dadhania D, Heyang M, Chen F, Suthanthiran M, Vlaminck IDe. Cell Free DNA Sequencing for Diagnosing Infections, Estimating Growth Rates, and Characterizing Antibiotic Resistomes in Urine Supernatants [abstract]. https://atcmeetingabstracts.com/abstract/cell-free-dna-sequencing-for-diagnosing-infections-estimating-growth-rates-and-characterizing-antibiotic-resistomes-in-urine-supernatants/. Accessed February 26, 2026.« Back to 2018 American Transplant Congress
